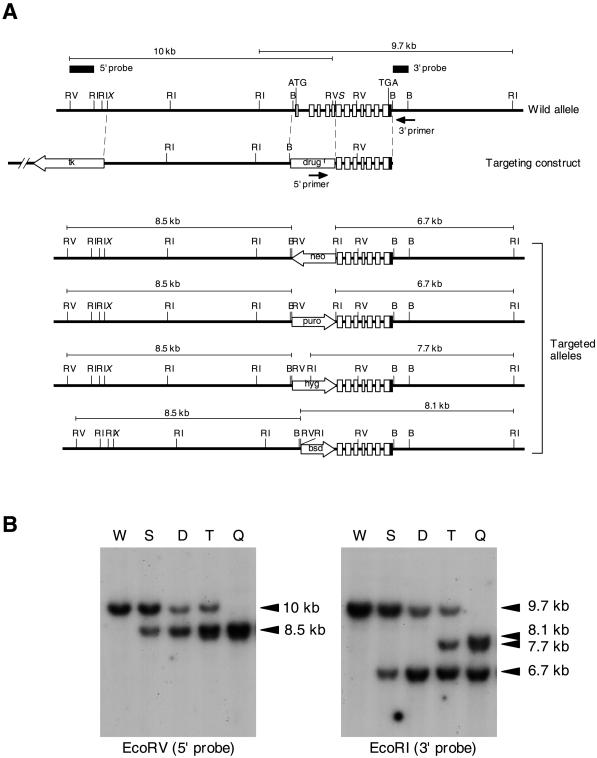
Figure 1.
Generation of mouse GAA1-knockout cells. (A) Structure of the mouse GAA1 gene (top), targeting constructs (middle), and expected disrupted alleles (bottom). Four consecutive recombination-targeting constructs in which drug resistance genes (drugr) were neomycin (neo), puromycin (puro), hygromycin (hyg), and blasticidin (bsd) resistance genes, respectively, were used in this order. The mouse GAA1 gene consists of 12 exons (shown in boxes: open areas, coding regions; closed areas, noncoding regions). Closed arrows represent PCR primers for screening recombinants. Probes for Southern analysis are indicated by thick bars. Predicted sizes of EcoRV- and EcoRI-digested fragments hybridized with 5′ and 3′ probes, respectively, are indicated above each allele. RV, EcoRV; RI, EcoRI; B, BamHI; X, XbaI; S, SacII (X and S are not unique). (B) Southern analysis of GAA1-targeted F9 cells. EcoRV-digested (left) and EcoRI-digested (right) genomic DNA from wild-type cells (W) and cells after single (S), double (D), triple (T), and quadruple (Q) recombinations were hybridized with the 5′ probe (left) and 3′ probe (right), respectively. The sizes of hybridized bands are indicated on the right.