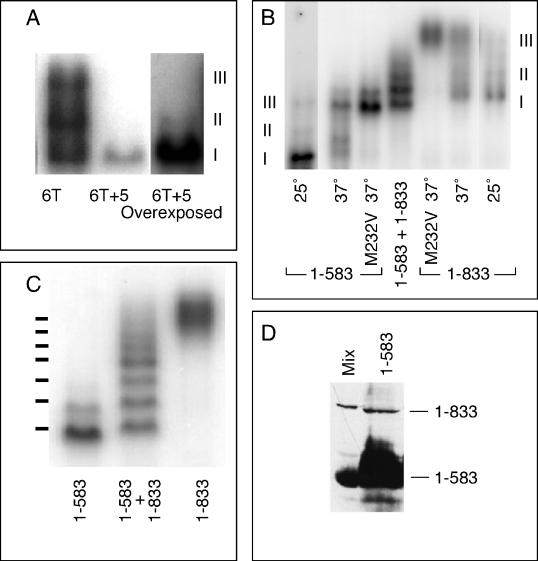
Figure 3.
Complex III is composed of two HSF trimers bound cooperatively to DNA. (A) DNA-binding reactions using as the probe BS6T (6T; left lane) or BS6T + 5, which differs from BS6T only in having 5 bp inserted between the first three and the last three AGAAG repeats (6T + 5). Because the 6T + 5 probe was of lower specific radioactivity, the right lane shows an overexposure to show the band of complex II. (B) DNA-binding reactions run with Tris-glycine buffer to identify the positions of complexes I, II, and III for HSF1–583 and HSF1–833. Nonshocked extracts show primarily complex I (lanes 1 [left lane] and 7 [right lane]), heat shock induces the formation of complex III (lanes 2 and 6), heat shock of the M232V mutant forms of these HSFs generates solely complex III (lanes 3 and 5), and coexpression of both long and short forms of HSF allows formation of heteromultimers (lane 4). (C) DNA-binding reactions with extracts from cells carrying the M232V versions of full-length HSF (1–833; right lane), HSF truncated at residue 583 by insertion of a TAA stop codon (1–583; left lane), or both (center lane) (a repeat of lanes 3–5 of B, but with the gel run for a longer time). The positions of the seven bands predicted if complex III is an HSF hexamer are shown to the left of the gel. (D) Western blot of a mixture of extracts from cells carrying full-length HSF or HSF1–583 (Mix) and of an extract from cells carrying only the plasmid-expressing HSF1–583 (1–583). The sample of HSF1–583 was overloaded to allow detection of the readthrough product.