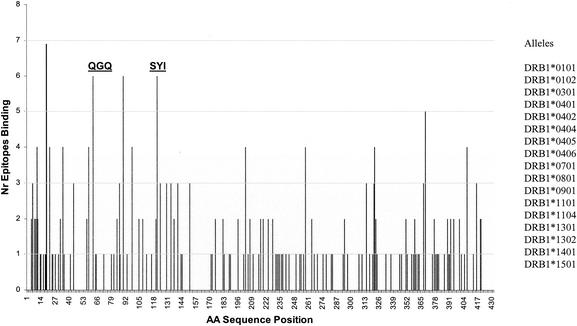
FIG. 1.
MHC class II motif prediction. A matrix-based MHC class II motif-matching algorithm was used to search the primary sequence of GbpB for known MHC class II binding motifs associated with MHC class II alleles. Shown is the result of the application of this algorithm for screening GbpB sequence against a set of 17 known MHC binding motifs (listed at right) associated with alleles at the MHC class II DRB1 locus. The ordinate represents the number (Nr) of motif matches associated with a given peak, while the abscissa represents the GbpB sequence position. The locations of the sequences used to make synthetic peptides for use in this study are indicated by “QGQ” and “SYI.” Regions of binding interest extend from 6 to 11 places to the right of peak residue 62 (associated with QGQ) and peak residue 121 (associated with SYI).