Table 1.
Data Collection, Phasing, and Refinement Statistics
| Native | Se (λ1) | Se (λ2) | Se (λ3) | |
|---|---|---|---|---|
| Diffraction Data | ||||
| Unit cell (Å) | a = 117.3, b = 117.3, c = 255.3 | a = 117.75, b = 117.75, c = 255.82 | ||
| Wavelength (Å) | 0.9797 | 0.97924 (peak) | 0.97939 (edge) | 0.97563 (remote) |
| Resolution (Å) | 2.85 | 3.1 | 3.1 | 3.1 |
| Space group | I41 | I4122 | I4122 | I4122 |
| No. of reflections | 117,971 | 52,416 | 52,519 | 52,339 |
| No. of unique reflections | 38,537 | 27,580 | 27,649 | 27,543 |
| Completeness | 96.2 (97.9) | 88.8 (91.0) | 88.6 (90.9) | 88.6 (90.9) |
| Multiplicity | 3.1 (3.0) | 1.9 | 1.9 | 1.9 |
| R-sym (%)a | 6.9 (43) | 13.5 (79.8) | 13.9 (79.8) | 13.1 (76.2) |
| I/σ(I) | 15.1 (2.2) | 7.1 (1.6) | 7.1 (1.6) | 7.6 (1.7) |
| Phasing | ||||
| Refined f′ (e-) | – | −8.56 | −10.3 | −5.34 |
| Refined f″ (e-) | – | 5.06 | 2.5 | 1.13 |
| Mean figure of merit | – | 0.36 | ||
| No. of Se sites | – | 10 | ||
| Structure Refinement | ||||
| Rfactor (%)b | 20.2 | – | – | – |
| Rfree (%) | 24.4 | – | – | – |
| No. of protein atoms | 7312 | – | – | – |
| No. of solvent atoms | 176 | – | – | – |
| No. of ligand atoms | 90 | – | – | – |
| RMS deviation of bond lengths (Å) | 0.007 | – | – | – |
| RMS deviation of bond angles (°) | 1.12 | – | – | – |
| RMS deviation of dihedral angles (°) | 9.35 | – | – | – |
| Average B factor (Å2) | 62.3 | – | – | – |
The numbers in parentheses indicate the value in the outer resolution shell. Rfree was calculated using 5% of reflections that were kept apart from the refinement during the whole process. RMS, root mean square.
R-sym =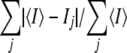 .
.
R-factor = 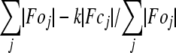 .
.
