Abstract
The phosphoprotein (P) of vesicular stomatitis virus (VSV) is a subunit of the viral RNA polymerase. In previous studies, we demonstrated that insertion of 19 amino acids in the hinge region of the protein had no significant effect on P protein function. In the present study, we inserted full-length enhanced green fluorescent protein (eGFP) in frame into the hinge region of P and show that the fusion protein (PeGFP) is functional in viral genome transcription and replication, albeit with reduced activity. A recombinant vesicular stomatitis virus encoding PeGFP in place of the P protein (VSV-PeGFP), which possessed reduced growth kinetics compared to the wild-type VSV, was recovered. Using the recombinant VSV-PeGFP, we show that the viral replication proteins and the de novo-synthesized RNA colocalize to sites throughout the cytoplasm, indicating that replication and transcription are not confined to any particular region of the cytoplasm. Real-time imaging of the cells infected with the eGFP-tagged virus revealed that, following synthesis, the nucleocapsids are transported toward the cell periphery via a microtubule (MT)-mediated process, and the nucleocapsids were seen to be closely associated with mitochondria. Treatment of cells with nocodazole or Colcemid, drugs known to inhibit MT polymerization, resulted in accumulation of the nucleocapsids around the nucleus and also led to inhibition of infectious-virus production. These findings are compatible with a model in which the progeny viral nucleocapsids are transported toward the cell periphery by MT and the transport may be facilitated by mitochondria.
Vesicular stomatitis virus (VSV) belongs to the family Rhabdoviridae in the order Mononegavirales. VSV is an enveloped virus with a negative-stranded RNA genome of 11,161 nucleotides. The genome encodes for five viral proteins, namely, nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and the RNA-dependent RNA polymerase (L) (1, 44). The viral genome is present within the virion as a ribonucleoprotein (RNP) being tightly encapsidated by the N protein and associated with the viral polymerase complex of L and P proteins (1, 44). The M protein is located underneath the viral envelope, whereas the G protein forms spikes on the viral envelope. The G protein binds to the cell surface receptor and is required for the entry of the virus into the cells. The virus enters susceptible cells by receptor-mediated endocytosis. Fusion of the viral envelope with the endosomal membrane in a pH-dependent process leads to the release of the viral nucleocapsids into the cytoplasm for viral gene expression. During VSV assembly, the progeny nucleocapsids are transported alone or in association with the viral M protein to the plasma membrane, where they are packaged into an envelope containing the viral G protein and are released from the infected cells. Although some of the fundamental steps in the VSV infection pathway have been deciphered, many questions concerning the entry mechanisms and transport of viral nucleocapsids to synthesis sites and from synthesis sites to assembly sites, as well as virus egress, remain poorly understood. Studies to provide an understanding of these processes have been hampered due to the inability to observe these events by live-cell imaging of infected cells.
In a recent report, it was demonstrated that the fusion of the viral envelope and the release of the nucleocapsid into the cytoplasm are two independent but successive steps in the endocytic pathway of VSV infection (29). These studies revealed that release of the viral nucleocapsid into the lumen of the endosomal vesicle occurs by the fusion of the viral envelope with the membranes of the endosomes but that the nucleocapsid release into the cytoplasm may require a back-fusion event in which the internal vesicles fuse with the membranes of the late endosome (29). Following synthesis in the cytoplasm, the progeny nucleocapsids must then be transported to the plasma membrane for viral assembly. The mechanism(s) by which the transport of such large RNP complexes is accomplished is unclear. Because of the high viscosity of the cytoplasm, movement of the RNPs by diffusion is likely to be limited (32). Intracellular pathogens and their macromolecular components overcome this obstacle by hijacking cytoplasmic motors and utilizing the cellular cytoskeleton as a roadway for intracellular transport to reach their destination (23, 35, 49, 50).
An understanding of some of the mechanistic details of virus entry by endocytic pathway and nucleocapsid release into the cytoplasm of infected cells has been possible only with the use of VSV chemically labeled with lipophilic fluorescent dyes (29). The use of such labeled viruses is limited to studies involving tracking of the input virus (16, 26, 29, 47). Once the viral nucleocapsid is delivered into the cytoplasm, subsequent tracking of the viral nucleocapsids, particularly, the sites of synthesis and transport of the nucleocapsids to the sites of virion assembly, would require genetic tagging of the viral nucleocapsid with fluorescent proteins.
VSV nucleocapsids are multiprotein-RNA complexes composed of viral RNA that is tightly wrapped with the N protein and is associated with P and L proteins. P protein is a multifunctional protein that is an essential subunit of the viral RNA-dependent RNA polymerase. In addition to its role in polymerase functions, it binds to the L protein and stabilizes it from proteolytic degradation (4, 13); it acts as a chaperone for the N protein, which then specifically encapsidates the viral RNA (9, 34, 41); and it interacts with terminal sequences of viral genome for viral RNA synthesis (21, 24). Our previous studies showed that the protein is organized in a modular fashion relative to its function (Fig. 1A) (6, 8, 19, 39). While phosphorylation of specific amino acid residues at the amino-terminal domain I (amino acid residues 1 to 150) is responsible for transcription activity (39), phosphorylation of specific amino acid residues at the carboxy-terminal domain II (amino acid residues 210 to 244) is important for optimal replication activity (19) of P protein. Phosphorylation of these residues at both domain I and domain II is indispensable for virus growth (6). Domain III, which comprises 21 to 25 residues at the extreme carboxy-terminal region, is important for mediating the binding of P protein to the N RNA template (8). The region that links domain I and domain II is called the hypervariable hinge region (approximately spanning amino acid residues 150 to 210). We recently studied the role of this hinge region and found that it plays an important role in VSV RNA synthesis and assembly of infectious particles (7). In that study, we also demonstrated that insertion of 19 amino acids (aa) within the hinge region of the protein (Fig. 1A) has no significant adverse effects on virus replication (7). In the present study, we show that a fusion protein (PeGFP), in which full-length enhanced green fluorescent protein (eGFP) was inserted in the hinge region of the P protein, is functional in viral genome transcription and replication. A recombinant VSV encoding the PeGFP protein in place of P protein (VSV-PeGFP) was recovered. Using VSV-PeGFP, we have examined the intracellular sites of viral RNA synthesis. By live-cell imaging of VSV-PeGFP-infected cells, we have found that the movement of newly synthesized viral nucleocapsids toward the cell periphery is mediated by microtubules (MTs). In addition, our studies indicate that mitochondria may play a role in intracellular transport of the viral nucleocapsids.
FIG. 1.
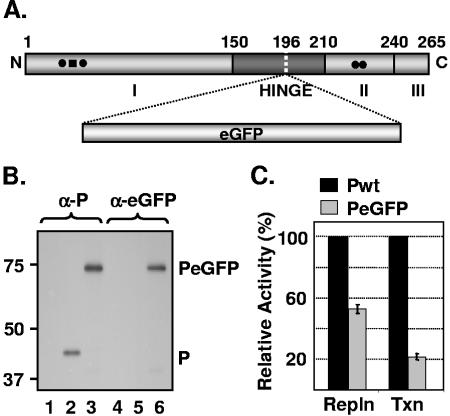
Replication and transcription activities of PeGFP fusion protein. (A) Domain organization of P protein, showing domains I, II, and III and the hinge region. The site of eGFP incorporation at aa position 196 is indicated by a vertical dotted line. (B) Expression of PeGFP fusion protein in transfected cells. Cells transfected with plasmids encoding P (lanes 2 and 5) or PeGFP (lanes 3 and 6) proteins or no plasmid (lanes 1 and 4) were radiolabeled with Expre35S35S label. The radiolabeled proteins were immunoprecipitated with antibodies as shown on the top (α-P, anti-P; α-eGFP, anti-eGFP), analyzed by SDS-PAGE, and detected by fluorography. Size markers in kDa are shown on the left. P and PeGFP proteins are identified on the right. (C) Replication and transcription activities of PeGFP protein relative to Pwt as determined by DI-particle replication or minigenome transcription assays (20, 40). The histograms represent the average data from three independent experiments, with standard deviations shown by error bars. Repln, replication; Txn, transcription.
MATERIALS AND METHODS
Cell culture, viruses, antibodies, and reagents.
Baby hamster kidney (BHK-21) cells were maintained as described before (6, 7). Propagation and preparation of stocks of VSV, recombinant vaccinia virus expressing T7 RNA polymerase (vTF7-3) (15), and defective-interfering (DI) particles were carried out in BHK-21 cells as described previously (40). Nocodazole (NOC), Colcemid, dimethyl sulfoxide (DMSO), 5-bromouridine 5′-triphosphate (BrUTP), and monoclonal antibody (MAb) against antitubulin (clone DM 1A) were from Sigma-Aldrich. NOC and Colcemid were reconstituted in DMSO; BrUTP was reconstituted in water. Rabbit polyclonal antibody against VSV P protein, mouse anti-VSV antibody, MAb against VSV N protein, and rabbit polyclonal antibody against the NH2-terminal region of VSV L protein have been previously described (6, 46). Rabbit polyclonal antibody against eGFP was prepared in the laboratory, and MAb against bromodeoxyuridine (BrdU) (clone BMC 9318) was obtained from Roche Diagnostics. Secondary antibodies, goat anti-mouse Alexa-488, goat anti-mouse Alexa-594, goat anti-rabbit Alexa-594, and MitoTracker Red were obtained from Molecular Probes; donkey anti-mouse Cy5 and donkey anti-rabbit Cy2 were from Jackson ImmunoResearch Laboratories.
Plasmid construction and virus recovery.
The construction of plasmids Pwt and PTn196 have been described in detail previously (7). Pwt is a plasmid encoding the wild-type (wt) VSV (VSVwt) (Indiana serotype) P protein in pGEM-3 vector (Promega) under the control of a T7 RNA polymerase promoter. The plasmid PTn196 is derived from Pwt by transposon-mediated mutagenesis and encodes the VSV P protein with a 19-aa insertion at residue 196. The eGFP sequence was amplified by PCR using primers eGFPNotF3, 5′ATATATGCGGCCGCAATGGTGAGCAAGGGC3′, and eGFPNotR3, 5′ATATATGCGGCCGCCTTGTACAGCTCGTC3′, each containing a NotI site (underlined); pIRES2eGFP (Invitrogen) as a template; and Pfu polymerase (Stratagene). The PCR product was digested with NotI and cloned at the unique NotI site in PTn196. The resulting plasmid, PTneGFP, encodes a fusion protein (PeGFP) of 524 amino acids, compared to wt P protein, which is 265 amino acids long.
The P protein coding region in the plasmid pVSVFL(+), which contains the full-length VSV genome (28), was replaced with the coding region for PeGFP from PTneGFP by use of the EcoRV sites that flank the P gene, as described previously (6). The resulting plasmid was designated pVSV-PeGFP, and the recombinant virus recovered from this plasmid was named VSV-PeGFP. For the generation of VSV encoding eGFP as an extra gene, an extra transcription unit was incorporated in the pVSVFL(+) plasmid between the G and L noncoding regions. The pVSVFL(+) plasmid was linearized with NheI, which is located between the G and L noncoding regions. To generate the extra transcription unit, two complementary oligonucleotides (FLVSVBsiWI+, 5′TATGAAAAAAACTAACAGATATCCGTACG3′, and FLVSVBsiWI−, 5′CGTACGGATAT CTGTTAGTTTTTTTCATA3′), incorporating a VSV poly(A)/termination signal, an intergenic dinucleotide, and a transcription initiation signal followed by a unique restriction site (BsiWI site is underlined), were designed. These oligonucleotides were annealed and kinased and then ligated with the linearized pVSVFL(+) plasmid, resulting in pVSVFLBsiWI. The eGFP coding region was amplified by PCR using the primers eGFPBsiWIF, 5′ATATATCGTACGGCCACCATGGTGAGCAAG3′, and eGFPBsiWIR, 5′ATATATCGTACGTTACTTGTACAGCTCGTC3′ (BsiWI site is underlined); digested with BsiWI; and cloned into pVSVFLBsiWI. Recombinant VSV recovered from the resulting plasmid, pVSV-eGFP, was named VSV-eGFP. The plasmids pN, pP, and pL (carrying the coding sequences of the N, P, and L proteins of VSV, respectively, and the plasmid p10BN), encoding a VSV minigenome, have been described previously (20, 38, 40). Recombinant viruses were recovered as described previously (6, 7).
Metabolic labeling and analysis of RNA.
Metabolic labeling and analysis of RNA in plasmid-transfected and virus-infected cells were performed as described previously (7, 20, 40). To examine transcription activity of mutant P proteins, BHK-21 cells were infected with vTF7-3 and subsequently transfected with p10BN, pN, and pL as well as Pwt or PTneGFP. RNAs were radiolabeled in the presence of actinomycin D and analyzed by electrophoresis in agarose-urea gel. To examine replication activity of mutant P proteins, cells infected with vTF7-3 and transfected with pN, pL, and Pwt or PTneGFP plasmids were superinfected with DI particles, and RNAs were radiolabeled and analyzed as described above. To examine the viral RNAs in infected cells, BHK-21 cells were infected with either wt VSV or eGFP-tagged viruses and the RNAs were radiolabeled and analyzed as described above.
Metabolic labeling and analysis of viral proteins.
Labeling of proteins in transfected or virus-infected cells, immunoprecipitation using anti-VSV antibody (1:200), anti-P antibody (1:200), and anti-eGFP antibody (1:200), sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, and detection of proteins by fluorography were carried out as described previously (6, 7). To examine proteins in virions, BHK-21 cells were infected with VSV at 1 PFU/cell and radiolabeled for 12 h at 4 h postinfection (hpi) with 100 μCi Expre35S35S per ml of 90% Dulbecco's modified Eagle's medium (DMEM) without methionine and cysteine and 10% regular DMEM. The virus from the clarified culture supernatant was pelleted through a 10% sucrose cushion at 38,000 rpm in a Beckman SW41 rotor for 1 h at 4°C. The virus pellets were resuspended, and viral proteins were analyzed by SDS-PAGE and detected as described above.
Determination of single-step growth kinetics.
Single-step growth kinetics of mutant and wt viruses were determined in BHK-21 cells essentially as described previously (6).
Drug treatment.
Cells were pretreated with 10 μg/ml NOC or Colcemid for 3 h at 37°C and infected with viruses for 45 min at 4°C. The cells were washed and incubated at 37°C in medium containing 10 μM NOC or Colcemid. As controls, NOC-untreated cells were maintained in media containing similar concentrations of DMSO.
BrUTP labeling of viral RNA.
BHK-21 cells were infected with VSV-PeGFP at a multiplicity of infection (MOI) of 10 PFU/cell and incubated for 2 h at 37°C. BrUTP labeling of de novo-synthesized viral RNA was performed as described previously (17), with minor modifications. Briefly, the infected cells were treated with 15 μg/ml of actinomycin D for 1 h at 2 hpi and transfected with BrUTP at a final concentration of 10 mM by use of Lipofectamine 2000 in the presence of actinomycin D for 2 h. The cells were fixed with 3.7% paraformaldehyde for 15 min at room temperature (RT) and processed for immunofluorescence for detection of viral RNA.
Fluorescence microscopy.
For epifluorescence and differential interference contrast (DIC) imaging, cells grown on glass coverslips or glass-bottomed cell culture dishes (MatTek) were either fixed as described above or visualized directly under an Olympus FV500/IX81 inverted confocal microscope. For immunofluorescence microscopy, the fixed cells were permeabilized with 0.1% Triton X-100 for 15 min at RT or with 100% ethanol at −20°C for 5 min. The cells were blocked with phosphate-buffered saline with 0.05% Tween 20 containing 3% bovine serum albumin for 30 min at RT. The primary and secondary antibodies diluted in phosphate-buffered saline with 0.05% Tween 20 containing 1% bovine serum albumin were added in succession after washing. The BrUTP-labeled RNAs were stained with an anti-BrdU MAb (1:10) followed by goat anti-mouse Alexa-594 (1:500) containing 1 U/μl of RNase inhibitor (RNase Out; Invitrogen). The cells were stained with DAPI (4′,6′-diamidino-2-phenylindole) and imaged using the inverted confocal microscope, and images were captured with a charge-coupled-device camera.
For real-time visualization of nucleocapsid movement, cells grown in glass-bottomed 35-mm dishes to 50% confluence were either treated with NOC or left untreated and then infected with VSV-PeGFP as described above and the dish was placed in a closed chamber maintained at 37°C and 5% CO2. Live-cell imaging was performed using the inverted confocal microscope fitted with a 100× lens. Images were captured at various times postinfection. For time-lapse recording of the same sets of cells, images were collected at intervals of 10 to 15 s for 30 min.
For staining of mitochondria, infected cells were washed thoroughly with serum-free DMEM containing 25 mM HEPES buffer. MitoTracker Red diluted in the above-described medium was added at 1 to 3 μM, and the cells were incubated at RT for 30 min. After being washed, the cells were maintained in medium containing serum and placed in the closed chamber.
RESULTS
PeGFP fusion protein supports transcription and replication of VSV.
Our recent observation (7) that insertion of 19 aa at position 196 in the hinge region of P protein had no significant adverse effect on transcription and replication prompted us to insert full-length eGFP coding sequence at this position (Fig. 1A). To determine if the insertion resulted in a stable protein, we examined the level of expression and the size of the PeGFP fusion protein in transfected cells. Accordingly, BHK-21 cells were transiently transfected with plasmids encoding either the wt or fusion proteins following vTF7-3 infection, and the proteins were radiolabeled and examined by immunoprecipitation with anti-P and/or anti-eGFP antibody and SDS-PAGE. As can be seen in Fig. 1B, the fusion protein PeGFP was stably expressed, could be immunoprecipitated with both anti-P and anti-eGFP antibodies, and possessed a predicted molecular mass of ∼73 kDa.
In order to examine the effect of eGFP insertion on P protein function in viral RNA synthesis, we used DI particles to determine replication activity and a minigenome template (p10BN) (20) to assess the transcription activity of PeGFP fusion protein. The radiolabeled DI RNAs and minigenome RNA were analyzed on urea-agarose gels and quantitated by densitometry. Results from three independent experiments show that the PeGFP protein is ∼55% active in replication, whereas it is ∼22% active in transcription compared to the wt P protein (Fig. 1C). These results suggest that the PeGFP fusion protein is functional, albeit with reduced activity.
Recovery of infectious VSV encoding PeGFP fusion protein.
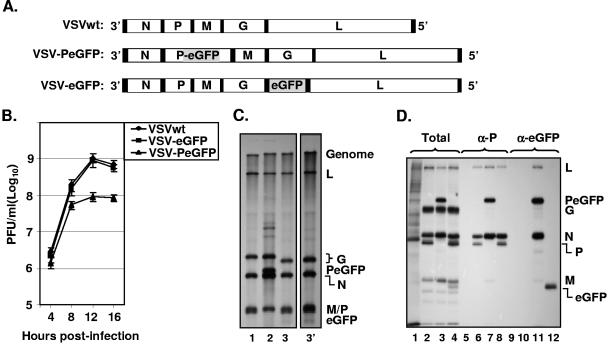
To test whether infectious virus could be recovered from VSV genome plasmids encoding P protein into which eGFP was inserted in frame in the hinge region, a recombinant VSV genome plasmid (pVSV-PeGFP) encoding PeGFP in place of wt P protein was constructed (Fig. 2A). We also constructed another recombinant VSV genome plasmid (pVSV-eGFP), containing eGFP coding sequence as an extra gene between the G and L gene junctions (Fig. 2A). Both of these genomic constructs led to recovery of recombinant VSV from transfected cells. In order to examine if the eGFP insertion into the P open reading frame has any effect on the growth of the virus, we examined single-cycle growth kinetics of the wt and mutant viruses. As can be seen from Fig. 2B, VSV-eGFP grew to titers similar to those of the wt VSV, whereas VSV-PeGFP grew to titers that were on average 8- to 10-fold less than those of the wt VSV. To determine if the viral growth correlated with the extent of viral macromolecular synthesis in infected cells, we examined RNA and viral protein synthesis in cells infected with these viruses. Our results show that overall synthesis of viral RNAs (Fig. 2C) and viral proteins (Fig. 2D) was not significantly different from results obtained with cells infected with wt VSV. These results indicate that in these mutant viruses the extent of viral protein and RNA synthesis did not correlate with the viral growth rate. The sizes of the viral mRNAs were as predicted, with the PeGFP mRNA migrating more slowly than mRNA for wt P protein or eGFP (Fig. 2C, compare lane 2 with lanes 1, 3, and 3′). As expected, six specific proteins (N, P, M, G, eGFP, and L) were detected in cells infected with VSV-eGFP (Fig. 2D, lane 4), whereas five proteins (N, PeGFP, M, G, and L) were detected in cells infected with VSV-PeGFP (Fig. 2D, lane 3). Immunoprecipitation of the proteins with anti-P (Fig. 2D, lanes 5 to 8) or anti-eGFP (Fig. 2D, lanes 9 to 12) antibody resulted in detection of P, PeGFP, and eGFP proteins of expected sizes from cells infected with the appropriate recombinant viruses. The N and L proteins and to some extent the M protein were also immunoprecipitated with these antibodies. This is not surprising since the P protein interacts with N and L proteins and is also associated with viral nucleocapsids that may contain M protein.
FIG. 2.
Recovery and characterization of recombinant VSV encoding PeGFP fusion protein. (A) Recombinant VSV genome plasmids. VSVwt, the wt VSV genome with the N, P, M, G, and L genes, shown in rectangular boxes; VSV-PeGFP, the VSV genome containing the PeGFP gene in place of the wt P gene; VSV-eGFP, the VSV genome containing the eGFP gene as an extra cistron. Intergenic regions as well as 3′ leader gene and 5′ trailer sequences are shown in black boxes; the eGFP coding region is shaded. (B) Single-cycle growth kinetics of mutant viruses. BHK-21 cells were infected with plaque-purified stocks of wt (VSVwt) or mutant (VSV-eGFP and VSV-PeGFP) viruses at an MOI of 20, and culture supernatants were collected at the indicated time points. The viruses in the supernatants were quantitated by plaque assay. The average values from four experiments are presented, with error bars representing standard deviations. (C) Analysis of VSV mRNAs in cells infected with the mutant viruses. BHK-21 cells were infected with VSVwt (lane 1), VSV-PeGFP (lane 2), and VSV-eGFP (lane 3) at an MOI of 10. Viral RNAs were radiolabeled, analyzed by electrophoresis, and detected by fluorography as described in Materials and Methods. Positions of the VSV mRNAs and full-length genome are indicated at the right. Lane 3′ shows a longer exposure of the autoradiogram to clearly identify the eGFP mRNA that is not readily visible in lane 3. (D) Analysis of proteins in cells infected with recombinant VSVs. BHK-21 cells were infected with VSVwt (lanes 2, 6, and 10), VSV-PeGFP (lanes 3, 7, and 11), and VSV-eGFP (lanes 4, 8, and 12) at an MOI of 10 or left uninfected (lanes 1, 5, and 9). The viral proteins were radiolabeled for 1 h at 4 hpi, analyzed by SDS-PAGE as total (lanes 1 to 4) or immunoprecipitated with anti-P (α-P) (lanes 5 to 8) or anti-eGFP (α-eGFP) (lanes 9 to 12) antibody, and detected by fluorography. The proteins are identified on the right.
Incorporation of reduced levels of PeGFP and L proteins into virions.
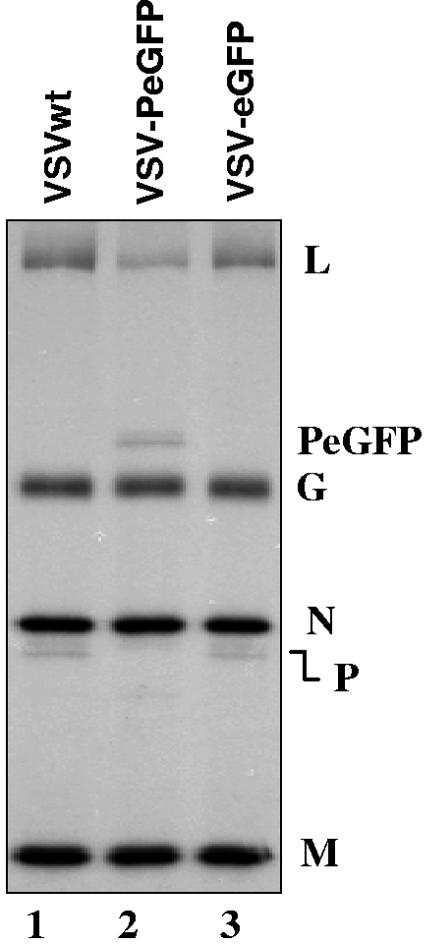
Since the overall size of PeGFP protein increased by almost twofold (524 aa for PeGFP as opposed to 265 aa for wt P protein), we were curious to examine what effect this would have on the efficiency of incorporation of PeGFP into VSV-PeGFP particles relative to other viral proteins as well as relative to wt P protein in VSV. To test the efficiency of incorporation of the PeGFP fusion protein into VSV-PeGFP particles, we radiolabeled the virus-infected cells with Expre35S35S, purified the extracellular virions, and analyzed the proteins incorporated into virions by SDS-PAGE analysis. Interestingly, we observed that PeGFP and L proteins were incorporated consistently at levels below that seen for wt VSV (Fig. 3). In these experiments, we examined viral proteins from all three viruses (VSVwt, VSV-PeGFP, and VSV-eGFP) that correspond to nearly equal amounts of N, M, and G proteins, the three major structural proteins within the virions. Unequivocally, the data shown in Fig. 3 and other similar independent experiments suggested that PeGFP and L proteins were incorporated less efficiently into VSV-PeGFP virions than into VSVwt and VSV-eGFP virions. Taking into account the number of methionine and cysteine residues in P and PeGFP proteins, we estimated from three independent experiments that the amounts of PeGFP and L proteins incorporated into the VSV-PeGFP virions were on average about 50% of the amounts of the P and L proteins in VSVwt. In the control virus expressing eGFP as an extra gene (VSV-eGFP), the incorporation of L and P proteins was similar to that of VSVwt. These results indicate that the larger PeGFP protein may have influenced incorporation of L-PeGFP protein complexes into the viral nucleocapsids that are packaged in the virions.
FIG. 3.
Incorporation of reduced levels of PeGFP and L proteins into VSV-PeGFP. Viral proteins in infected cells were radiolabeled, and the proteins incorporated into purified virions were analyzed by SDS-PAGE and detected as described in the text. The positions of various proteins are shown on the right.
Examination of intracellular sites of viral RNA synthesis.
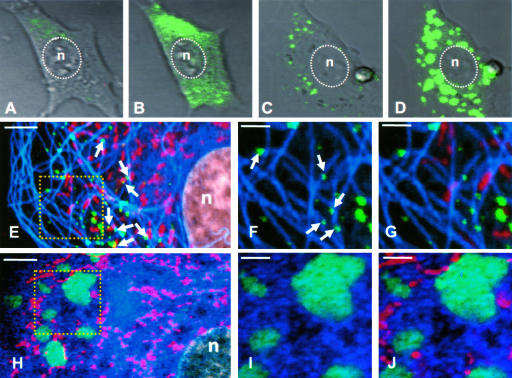
Subcellular localizations of eGFP were noticeably different in cells infected with VSV-eGFP and VSV-PeGFP. While VSV-eGFP-infected cells showed typical eGFP distribution evenly throughout the cytoplasm as well as in the nucleus (Fig. 4A), the fluorescence pattern in cells infected with VSV-PeGFP appeared granular and the granules were distributed throughout the cytoplasm but not in the nucleus (Fig. 4B). The differential distribution patterns of PeGFP and eGFP likely reflect the fact that PeGFP is associated with the viral nucleocapsids and is intimately involved in viral genome replication and transcription, whereas eGFP is not.
FIG. 4.
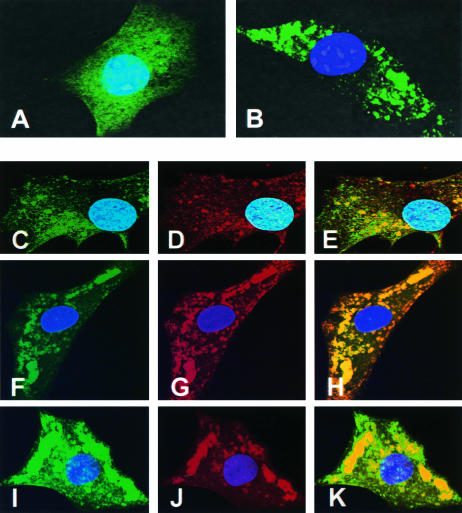
Examination of sites of viral RNA synthesis in cells infected with VSV-PeGFP. (A and B) Distribution of fluorescence in cells infected with recombinant VSVs. BHK-21 cells infected with VSV-eGFP (A) or VSV-PeGFP (B) at an MOI of 10 were fixed at 4 hpi, stained with DAPI, and examined by fluorescence microscopy. (C to E) Cells infected with VSV-PeGFP were labeled with BrUTP, and the de novo-synthesized RNA was detected by MAb to BrdU and goat anti-mouse Alexa-594. Colocalization of PeGFP (C) with RNA (D) is shown in the merged image (E). (F to K) VSV-PeGFP-infected cells were fixed at 4 hpi and stained with anti-N MAb (F to H) or anti-L antibody (I to K) and the corresponding secondary antibodies conjugated to Alexa-594. Colocalization of N (G) or L (J) with PeGFP (F and I) is shown in the merged images (H and K).
To determine if the locations of the granular fluorescence seen in cells infected with VSV-PeGFP represent the sites of synthesis of viral RNA, we examined the sites of de novo synthesis of viral RNA. Accordingly, cells infected with VSV-PeGFP were treated with actinomycin D followed by BrUTP to label de novo-synthesized RNA and examined by immunofluorescence using anti-BrdU antibody. Newly synthesized RNA labeled with BrUTP was detected throughout the cytoplasm (Fig. 4D), and PeGFP protein (Fig. 4C) colocalized to these sites (Fig. 4E). The viral N and L proteins also colocalized with the PeGFP protein (Fig. 4F to K) in a manner similar to viral RNA. These data suggest that the de novo-synthesized RNAs colocalize with the viral N, L, and PeGFP proteins, indicating that synthesis of viral RNA occurs at these sites. The sites of viral RNA synthesis appear to be distributed throughout the cytoplasm. The staining observed near the cell periphery may represent the viral nucleocapsids in transit to the cell surface for assembly.
Association of viral nucleocapsids with mitochondria.
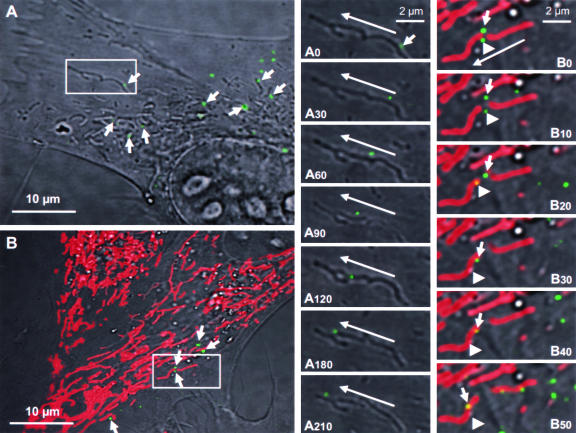
Live-cell imaging of VSV-PeGFP-infected cells revealed that many green fluorescent dots were detectable as early as 30 min after infection and became numerous by 1 to 2 hpi (data not shown). A size estimation of several individual green dots suggested that they represent individual viral nucleocapsids. Most of these fluorescent nucleocapsids were mobile within the cytoplasm with time, moving toward the cell periphery away from the nucleus. High-magnification DIC images of infected cells showed that many of these nucleocapsids (Fig. 5A) appeared to be moving along with or on mitochondrion-like structures. Time-lapse imaging of one such nucleocapsid (Fig. 5A) indicated that the fluorescent nucleocapsids traversed along or in close association with mitochondria or mitochondrion-like structures in a nonlinear fashion toward the cell periphery (Fig. 5A0 to A210). Tracking of several such nucleocapsids (n = 12) over time indicated that they moved with an average speed of approximately 30 nm/s.
FIG. 5.
Live-cell tracking of VSV fluorescent nucleocapsid movement in infected cells. (A) BHK-21 cells were infected with VSV-PeGFP at an MOI of 10, and at 2 hpi, the culture dish was transferred to a 37°C chamber with 5% CO2 and observed under an inverted laser scanning microscope. A single infected cell and a small area (rectangular box) containing one fluorescent nucleocapsid were observed with time. Arrows in this panel identify some nucleocapsids that are in close association with mitochondrion-like structures. Panels A0 to A210 are close-up images of the small area showing the movement of a nucleocapsid (small arrow in A0) with time from the beginning (A0) to 30 s (A30), 60 s (A60), 90 s (A90), 120 s (A120), 180 s (A180), and 210 s (A210) of image recording. The direction of movement of the nucleocapsid (long arrows) toward the cell periphery is shown. (B) Live-cell tracking of nucleocapsids in infected cells stained with MitoTracker Red, which specifically stains mitochondria. The experiment was performed as described for panel A except that the infected cells were treated with MitoTracker Red for 30 min prior to image recording. Arrows identify some nucleocapsids that are in close association with red-stained mitochondria. Panels B0 (beginning) to B50 (50 s) are close-up images of the area boxed in panel B and represent the images recorded at times in seconds, as described for panels A0 to A210. Two fluorescent nucleocapsids (identified by small arrow and arrowhead) are seen moving in close association with mitochondria (red). The long arrow shows the direction of movement of the nucleocapsids toward the cell periphery.
To confirm that the viral nucleocapsids were moving along and/or associated with mitochondria, we examined the infected cells treated with MitoTracker Red, which specifically stains only mitochondria. By live-cell imaging, the green fluorescent nucleocapsids were seen closely associated with the red-stained mitochondria (Fig. 5B) and were moving along mitochondria with time (Fig. 5B0 to B50). Tracking of two such nucleocapsids (Fig. 5B0 to B50) indicates that they are clearly seen very close to, if not attached to, mitochondria. These results suggest that mitochondria may be involved in transport of viral nucleocapsids from the sites of synthesis to the cell periphery.
Involvement of MTs in transport of progeny viral nucleocapsids.
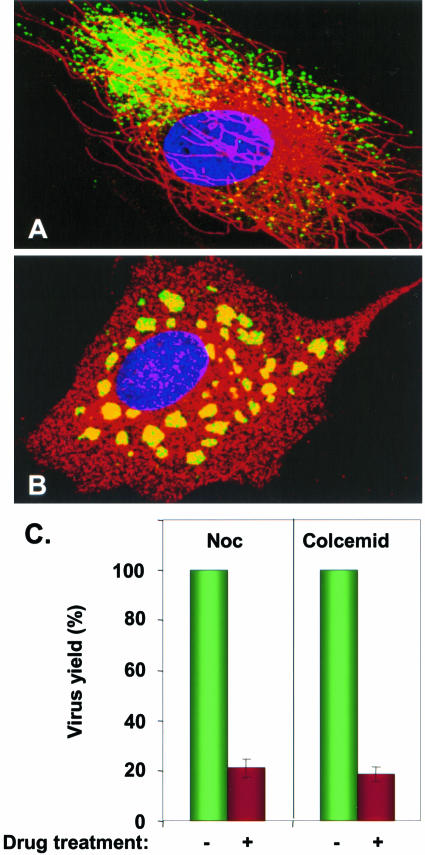
Since it is known that movement and intracellular distribution of mitochondria are dependent on cytoskeletal components, especially the MTs (18, 54), we investigated whether MTs are involved in the observed movement of the viral nucleocapsids. Initially, we examined the distribution of fluorescent viral nucleocapsids in infected cells in the absence or presence of NOC, a drug known to inhibit microtubule polymerization. Cells not treated with the drug and infected with VSV-PeGFP showed typical MT distribution in the cytoplasm (Fig. 6A). In these cells, the fluorescent viral nucleocapsids appeared to be concentrated on one side of the nucleus, although nucleocapsids were also seen distributed throughout the cytoplasm (Fig. 6A). This was representative of most infected cells. In addition, high-magnification images revealed that many of these nucleocapsids were found to be closely associated with MTs. Cells pretreated with NOC and infected with VSV-PeGFP with continued presence of the drug showed complete loss of MTs, and a different pattern of distribution of viral nucleocapsids was observed (Fig. 6B). The vast majority of the viral nucleocapsids were seen clustered around the nucleus as aggregates (Fig. 6B), although a small number of individual nucleocapsids were also seen. These data suggest that, in the absence of MTs, the majority of the nucleocapsids are localized around the nucleus and fail to be transported from these sites. A similar distribution of viral nucleocapsids was observed with Colcemid (data not shown), which also inhibits microtubule polymerization. The overall yield of extracellular virus from cells treated with NOC or Colcemid was about 20% of that obtained from control untreated cells (Fig. 6C). The effect of the drugs on virus yield was not due to reduced levels of viral protein synthesis or genome replication (data not shown).
FIG. 6.
Role of MTs in nucleocapsid transport. Untreated (A) or NOC-treated (B) cells were infected with VSV-PeGFP, fixed at 4 hpi, and stained with antitubulin antibody. MTs are stained red, while the nucleocapsids appear in green. The nucleus stained with DAPI is shown in blue. (C) Effect of NOC and Colcemid on virus yield. Cells were either treated (+) with NOC or Colcemid or left untreated (−) and then infected with VSV-PeGFP at an MOI of 10. The cell culture supernatant was harvested at 12 to 14 hpi, and the virus yield was determined by plaque assay. The average data from three independent experiments are presented, with standard deviations shown by error bars.
A time course experiment of untreated and NOC-treated cells infected with VSV-PeGFP was performed to examine, by live-cell recording, the synthesis of progeny nucleocapsids for up to 4 h. Different patterns of fluorescence distribution were observed in cells without and with NOC treatment. At the beginning of infection, green fluorescence was barely detectable, but at 1 hpi, fluorescence was readily detectable in untreated and drug-treated cells and gradually increased in intensity with time. By 2 and 4 hpi, numerous nucleocapsids were visible in the cytoplasm of untreated cells (Fig. 7A and B). In contrast, nucleocapsids were seen in aggregates at 2 hpi (Fig. 7C) and became larger, brighter, and more numerous by 4 hpi (Fig. 7D).
FIG. 7.
Imaging of VSV-PeGFP-infected cells to demonstrate the effect on NOC on nucleocapsid transport. (A to D) DIC images of live cells showing fluorescent nucleocapsid synthesis in infected cells at 2 (A and C) or 4 (B and D) hpi without (A and B) or with (C and D) NOC treatment. The nucleus (n) is marked by a dotted oval. (E to G) Untreated cells infected with VSV-PeGFP were stained with MitoTracker Red at 4 hpi, fixed, and immunostained with antitubulin antibody. VSV nucleocapsids (green), mitochondria (red), and MTs (pseudocolor blue) were visualized by confocal fluorescence microscopy. The area in the square in panel E is magnified in panels F and G. Arrows in panel E show some nucleocapsids in association with mitochondria. Arrows in panel F show some nucleocapsids directly on MT tracks. (H to J) Same as in panels E to G, but with NOC treatment. The area in the square in panel H is magnified in panels I and J. Bars, 5 μm (E and H) and 2 μm (F, G, I, and J).
Immunostaining of infected cells with antitubulin antibody in the presence of MitoTracker Red and with or without NOC treatment further confirmed the involvement of MTs in the transport of nucleocapsids. In the absence of NOC (Fig. 7E), the nucleocapsids were dispersed throughout the cytoplasm and largely remained as individual nucleocapsids, the majority of which were associated with or in close proximity to MTs as well as mitochondria (Fig. 7F and G). In cells treated with NOC (Fig. 7H to J), aggregates of fluorescent nucleocapsids were detected. Thus, the loss of MTs in NOC-treated cells led to an accumulation of progeny viral nucleocapsids in the cytoplasm, resulting in the formation of these aggregates.
DISCUSSION
In recent years, the use of fluorescent molecules as protein tags, combined with advanced imaging techniques, has been instrumental in providing unprecedented opportunities for visualizing many dynamic processes in living cells (12, 27, 31). By use of viruses genetically tagged with fluorescent proteins, several aspects of virus biology and virus-cell interactions in vitro and in vivo have been examined. In particular, fluorescent-tagged viruses were used to study intracellular transport of human immunodeficiency virus capsids (35), capsid assembly (36), and genome recombination (30, 43); herpes virus entry, capsid transport, and egress (33, 45, 48); rhabdovirus gene expression (14) and immune response (25); and morbillivirus tissue tropism (53). In this report, we generated a recombinant VSV encoding P protein fused in frame with eGFP (VSV-PeGFP), which allowed us to track the movement of viral nucleocapsids in infected cells by real-time imaging. Our results suggest that the nucleocapsids are synthesized at sites distributed throughout the cytoplasm and that the progeny nucleocapsids are transported to the cell periphery by an MT-mediated process. Our observation that nucleocapsids are in close proximity to mitochondria suggests that mitochondria may also be involved in the nucleocapsid transport process.
Insertion of eGFP into the hinge region of the P protein led to PeGFP fusion protein, which was active in viral genome transcription and replication, indicating that the hinge region can accommodate large insertions without much adverse effect on protein function. However, incorporation of PeGFP as well as the L protein into virions was affected (Fig. 3). It is possible that the large size of the fusion protein might have sterically hindered the association of P-L complexes with the viral N RNA template, resulting in reduced incorporation of PeGFP and L proteins in the virions. Although the levels of viral macromolecular synthesis in cells infected with VSV, VSV-PeGFP, or VSV-GFP were not significantly different from each other, VSV-PeGFP grew to titers that were almost 10-fold less than those of the other viruses. Whether the reduced levels of incorporation of L and P proteins in VSV-PeGFP account for its retarded growth phenotype remains to be examined. We have recently demonstrated that the P protein of VSV plays a major role in assembly of infectious particles (7). It is therefore possible that insertion of eGFP into P protein may have affected the assembly functions of the protein to some extent, resulting in retarded growth of VSV-PeGFP. It should be noted that a recombinant rabies virus encoding the P protein with eGFP fused in frame at its amino terminus possessed a significantly retarded growth phenotype (14). In addition, eGFP incorporation into the L protein of measles virus (11) led to a reduced growth phenotype of the recombinant virus. However, a recombinant rinderpest virus with insertion of eGFP at a similar location in the L protein grew like the wt virus in vitro but was attenuated for growth and virulence in its natural animal host, cattle (2). Clearly, these data indicate that growth and virulence properties can be altered by insertion of eGFP into the viral polymerase. It will be interesting to examine whether VSV-PeGFP with insertion of eGFP in the P protein of the viral polymerase complex possesses altered growth and virulence phenotypes in vivo.
In cells infected with VSV-PeGFP, the intracellular distribution of fluorescence was punctate. The majority of these puncta were seen distributed throughout the cytoplasm, and their locations represented the sites of viral replication since the de novo-synthesized RNA as well as other components of viral replication machinery, namely, the N and the L proteins, colocalized to these sites. The interpretation that viral RNA synthesis occurs throughout the cytoplasm but in proximity to the cell nucleus was strengthened by the observation that in cells treated with NOC or Colcemid, which inhibit MT polymerization, leading to a disruption of nucleocapsid transport from the sites of synthesis to the cell periphery, the nucleocapsids were seen as aggregates around the nucleus (Fig. 6 and 7).
VSV contains approximately 450 copies of P molecules per nucleocapsid (51). Although VSV-PeGFP contains almost half as many PeGFP molecules as there are P molecules in wt VSV (Fig. 3), it still has more than the 120 copies of eGFP necessary to detect individual rotavirus particles (5). Clearly, by live-cell imaging of infected cells at high magnification, we were able to detect individual nucleocapsids as tiny green fluorescent structures (Fig. 5 and 6). Many of these green structures were of uniform size, roughly approximating the size of viral nucleocapsids, although larger green dots, presumably representing multiple nucleocapsids in very close proximity, were also observed in drug-untreated cells. These green structures were not seen in cells expressing PeGFP alone or in combination with N and/or L proteins (data not shown). Many of these fluorescent nucleocapsids appeared to be associated with mitochondria and moved in parallel with the longitudinal axis of mitochondria toward the cell periphery. The significance of the association of the nucleocapsid with mitochondria is not clear at this time, but it is possible that cytoskeletal structures, like MTs, that are used by mitochondria for their intracellular distribution (18, 54) might be involved in nucleocapsid transport. The association of nucleocapsids with mitochondria may thus be transient and possibly serve as bridges linking MTs during transport of the nucleocapsids. Alternatively, it is possible that the association of nucleocapsids with mitochondria may be just random. Further work will be necessary to provide any functional significance to the association of nucleocapsids with mitochondria.
Although the effect of NOC on nucleocapsid distribution was dramatic (Fig. 6B), virus yield was reduced to about 20% of that from untreated control cells (Fig. 6C). It is possible that nucleocapsid transport may occur by additional mechanisms that are independent of MT. In this regard, it is of note that for DNA viruses whose capsids are specifically transported by MTs toward the cell periphery, a 20 to 25% reduction in virus yield in the presence of NOC has been considered to be significant (3, 37). In the light of these observations, our data are consistent with the interpretation that MTs are involved in the transport of viral nucleocapsids by an anterograde movement toward the cell periphery. Although the nucleocapsid transport toward the cell periphery was affected by NOC or Colcemid treatment, virus entry and uncoating remained unaffected, as seen by synthesis and accumulation of viral nucleocapsids with time in NOC-treated cells (Fig. 7C and D). This is consistent with the recent findings that NOC has no significant adverse effect on VSV entry, uncoating, or viral macromolecular synthesis (29).
How might the MTs be involved in transport of viral nucleocapsids toward the cell periphery? Since the MTs form tracks on which cellular cargos are transported by intracellular kinesin and dynein motors (10, 52), it is possible to envision a scenario in which the nucleocapsids interact with kinesin and/or dynein motors and are transported by these motors on MT tracks. In this regard, it is interesting to note that the P protein of rabies virus (another rhabdovirus) has been shown to interact efficiently with the dynein light chain (14, 22, 42), a component of the dynein motor. Although the significance of this interaction in terms of intracellular transport of the viral nucleocapsids has not been established, it is tempting to speculate that VSV nucleocapsids also interact with one or more components of the intracellular motors that play a role in the transport of the nucleocapsids mediated by MTs. With the availability of fluorescently tagged nucleocapsids, it will be possible to study these interactions and examine intracellular events by real-time imaging of virus-infected cells for a better understanding of the mechanisms by which the viral nucleocapsids are transported from the sites of synthesis to the assembly sites. Furthermore, VSV-PeGFP could be used to study entry and uncoating mechanisms that may provide significant insight into receptor-mediated endocytosis and uncoating of viral nucleocapsids in infected cells.
Acknowledgments
We thank A. Martinsen and T. Fangman for excellent technical help.
This investigation was supported by a grant (AI 34956) from NIAID, NIH, and also in part by P20RR15635 from NCRR, NIH.
REFERENCES
- 1.Banerjee, A. K. 1987. Transcription and replication of rhabdoviruses. Microbiol. Rev. 51:66-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, D. D., B. K. Rima, I. V. Allen, M. D. Baron, A. C. Banyard, T. Barrett, and W. P. Duprex. 2005. Rational attenuation of a morbillivirus by modulating the activity of the RNA-dependent RNA polymerase. J. Virol. 79:14330-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukrinskaya, A., B. Brichacek, A. Mann, and M. Stevenson. 1998. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J. Exp. Med. 188:2113-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canter, D. M., and J. Perrault. 1996. Stabilization of vesicular stomatitis virus L polymerase protein by P protein binding: a small deletion in the C-terminal domain of L abrogates binding. Virology 219:376-386. [DOI] [PubMed] [Google Scholar]
- 5.Charpilienne, A., M. Nejmeddine, M. Berois, N. Parez, E. Neumann, E. Hewat, G. Trugnan, and J. Cohen. 2001. Individual rotavirus-like particles containing 120 molecules of fluorescent protein are visible in living cells. J. Biol. Chem. 276:29361-29367. [DOI] [PubMed] [Google Scholar]
- 6.Das, S. C., and A. K. Pattnaik. 2004. Phosphorylation of vesicular stomatitis virus phosphoprotein P is indispensable for virus growth. J. Virol. 78:6420-6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das, S. C., and A. K. Pattnaik. 2005. Role of the hypervariable hinge region of phosphoprotein P of vesicular stomatitis virus in viral RNA synthesis and assembly of infectious virus particles. J. Virol. 79:8101-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das, T., A. K. Pattnaik, A. M. Takacs, T. Li, L. N. Hwang, and A. K. Banerjee. 1997. Basic amino acid residues at the carboxy-terminal eleven amino acid region of the phosphoprotein (P) are required for transcription but not for replication of vesicular stomatitis virus genome RNA. Virology 238:103-114. [DOI] [PubMed] [Google Scholar]
- 9.Davis, N. L., H. Arnheiter, and G. W. Wertz. 1986. Vesicular stomatitis virus N and NS proteins form multiple complexes. J. Virol. 59:751-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dohner, K., C. H. Nagel, and B. Sodeik. 2005. Viral stop-and-go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol. 13:320-327. [DOI] [PubMed] [Google Scholar]
- 11.Duprex, W. P., F. M. Collins, and B. K. Rima. 2002. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame. J. Virol. 76:7322-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrhardt, D. 2003. GFP technology for live cell imaging. Curr. Opin. Plant Biol. 6:622-628. [DOI] [PubMed] [Google Scholar]
- 13.Emerson, S. U., and M. Schubert. 1987. Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 84:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finke, S., K. Brzozka, and K. K. Conzelmann. 2004. Tracking fluorescence-labeled rabies virus: enhanced green fluorescent protein-tagged phosphoprotein P supports virus gene expression and formation of infectious particles. J. Virol. 78:12333-12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgi, A., C. Mottola-Hartshorn, A. Warner, B. Fields, and L. B. Chen. 1990. Detection of individual fluorescently labeled reovirions in living cells. Proc. Natl. Acad. Sci. USA 87:6579-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heggeness, M. H., M. Simon, and S. J. Singer. 1978. Association of mitochondria with microtubules in cultured cells. Proc. Natl. Acad. Sci. USA 75:3863-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang, L. N., N. Englund, T. Das, A. K. Banerjee, and A. K. Pattnaik. 1999. Optimal replication activity of vesicular stomatitis virus RNA polymerase requires phosphorylation of a residue(s) at carboxy-terminal domain II of its accessory subunit, phosphoprotein P. J. Virol. 73:5613-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang, L. N., N. Englund, and A. K. Pattnaik. 1998. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions. J. Virol. 72:1805-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isaac, C. L., and J. D. Keene. 1982. RNA polymerase-associated interactions near template promoter sequences of defective interfering particles of vesicular stomatitis virus. J. Virol. 43:241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacob, Y., H. Badrane, P. E. Ceccaldi, and N. Tordo. 2000. Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J. Virol. 74:10217-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jouvenet, N., P. Monaghan, M. Way, and T. Wileman. 2004. Transport of African swine fever virus from assembly sites to the plasma membrane is dependent on microtubules and conventional kinesin. J. Virol. 78:7990-8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keene, J. D., B. J. Thornton, and S. U. Emerson. 1981. Sequence-specific contacts between the RNA polymerase of vesicular stomatitis virus and the leader RNA gene. Proc. Natl. Acad. Sci. USA 78:6191-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koser, M. L., J. P. McGettigan, G. S. Tan, M. E. Smith, H. Koprowski, B. Dietzschold, and M. J. Schnell. 2004. Rabies virus nucleoprotein as a carrier for foreign antigens. Proc. Natl. Acad. Sci. USA 101:9405-9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakadamyali, M., M. J. Rust, H. P. Babcock, and X. Zhuang. 2003. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. USA 100:9280-9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lalonde, S., D. W. Ehrhardt, and W. B. Frommer. 2005. Shining light on signaling and metabolic networks by genetically encoded biosensors. Curr. Opin. Plant Biol. 8:574-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawson, N. D., E. A. Stillman, M. A. Whitt, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 92:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Blanc, I., P. P. Luyet, V. Pons, C. Ferguson, N. Emans, A. Petiot, N. Mayran, N. Demaurex, J. Faure, R. Sadoul, R. G. Parton, and J. Gruenberg. 2005. Endosome-to-cytosol transport of viral nucleocapsids. Nat. Cell Biol. 7:653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy, D. N., G. M. Aldrovandi, O. Kutsch, and G. M. Shaw. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl. Acad. Sci. USA 101:4204-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lippincott-Schwartz, J., and G. H. Patterson. 2003. Development and use of fluorescent protein markers in living cells. Science 300:87-91. [DOI] [PubMed] [Google Scholar]
- 32.Luby-Phelps, K. 2000. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int. Rev. Cytol. 192:189-221. [DOI] [PubMed] [Google Scholar]
- 33.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masters, P. S., and A. K. Banerjee. 1988. Resolution of multiple complexes of phosphoprotein NS with nucleocapsid protein N of vesicular stomatitis virus. J. Virol. 62:2651-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller, B., J. Daecke, O. T. Fackler, M. T. Dittmar, H. Zentgraf, and H. G. Krausslich. 2004. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J. Virol. 78:10803-10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naranatt, P. P., H. H. Krishnan, M. S. Smith, and B. Chandran. 2005. Kaposi's sarcoma-associated herpesvirus modulates microtubule dynamics via RhoA-GTP-diaphanous 2 signaling and utilizes the dynein motors to deliver its DNA to the nucleus. J. Virol. 79:1191-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pattnaik, A. K., L. A. Ball, A. W. LeGrone, and G. W. Wertz. 1992. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell 69:1011-1020. [DOI] [PubMed] [Google Scholar]
- 39.Pattnaik, A. K., L. Hwang, T. Li, N. Englund, M. Mathur, T. Das, and A. K. Banerjee. 1997. Phosphorylation within the amino-terminal acidic domain I of the phosphoprotein of vesicular stomatitis virus is required for transcription but not for replication. J. Virol. 71:8167-8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pattnaik, A. K., and G. W. Wertz. 1990. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J. Virol. 64:2948-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peluso, R. W. 1988. Kinetic, quantitative, and functional analysis of multiple forms of the vesicular stomatitis virus nucleocapsid protein in infected cells. J. Virol. 62:2799-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raux, H., A. Flamand, and D. Blondel. 2000. Interaction of the rabies virus P protein with the LC8 dynein light chain. J. Virol. 74:10212-10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhodes, T. D., O. Nikolaitchik, J. Chen, D. Powell, and W. S. Hu. 2005. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J. Virol. 79:1666-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose, J. K., and M. A. Whitt. 2001. Rhabdoviridae: the viruses and their replication, p. 1221-1244. In B. N. Fields and D. M. Knipe (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 45.Sampaio, K. L., Y. Cavignac, Y. D. Stierhof, and C. Sinzger. 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J. Virol. 79:2754-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schubert, M., G. G. Harmison, C. D. Richardson, and E. Meier. 1985. Expression of a cDNA encoding a functional 241-kilodalton vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. USA 82:7984-7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seisenberger, G., M. U. Ried, T. Endress, H. Buning, M. Hallek, and C. Brauchle. 2001. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science 294:1929-1932. [DOI] [PubMed] [Google Scholar]
- 48.Smith, G. A., L. Pomeranz, S. P. Gross, and L. W. Enquist. 2004. Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc. Natl. Acad. Sci. USA 101:16034-16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suomalainen, M., M. Y. Nakano, S. Keller, K. Boucke, R. P. Stidwill, and U. F. Greber. 1999. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 144:657-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas, D., W. W. Newcomb, J. C. Brown, J. S. Wall, J. F. Hainfeld, B. L. Trus, and A. C. Steven. 1985. Mass and molecular composition of vesicular stomatitis virus: a scanning transmission electron microscopy analysis. J. Virol. 54:598-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vale, R. D. 2003. The molecular motor toolbox for intracellular transport. Cell 112:467-480. [DOI] [PubMed] [Google Scholar]
- 53.von Messling, V., D. Milosevic, and R. Cattaneo. 2004. Tropism illuminated: lymphocyte-based pathways blazed by lethal morbillivirus through the host immune system. Proc. Natl. Acad. Sci. USA 101:14216-14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yaffe, M. P., N. Stuurman, and R. D. Vale. 2003. Mitochondrial positioning in fission yeast is driven by association with dynamic microtubules and mitotic spindle poles. Proc. Natl. Acad. Sci. USA 100:11424-11428. [DOI] [PMC free article] [PubMed] [Google Scholar]