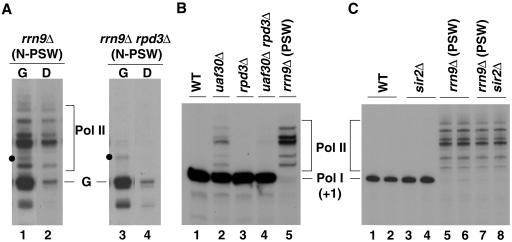
FIG. 3.
rpd3Δ mutation inhibits Pol II transcription of rRNA genes, but not Pol I transcription of rRNA genes or Pol II transcription of GAL7-35S rRNA gene fusion gene. (A) The two strains, NOY2111 (rrn9Δ, N-PSW) and its rpd3Δ derivative (Fig. 2A), were grown in YEPG complete medium to a cell density of A600 of ∼0.2. Cells from half of each culture were collected by centrifugation, resuspended in YEPD complete medium, and incubated for 1 h (D lanes). The remaining half of each culture was kept in YEPG complete medium and incubated as controls (G lanes). Total cellular RNA was then isolated from each culture. RNA transcribed from the GAL7-35S rRNA gene fusion gene (the main band labeled G) and RNA derived from chromosomal rRNA genes by Pol II transcription (bands labeled Pol II) were analyzed by primer extension using 2.5 μg RNA. Relevant lanes from an autoradiogram of a single gel are shown. It should be noted that there are some bands (e.g., those indicated with a dot) above the main transcription start site (G) of the GAL7-35S rRNA gene fusion gene that, like band G, were strongly reduced upon transfer to glucose and therefore, almost certainly represent transcripts from the GAL7 promoter. Quantification of radioactive bands indicated that rRNA synthesized from the GAL7 promoter in galactose was comparable in the two strains (the amount of band G in lane 3/amount of band G in lane 1 = 0.78), whereas Pol II transcription of chromosomal rRNA genes in the rrn9Δ rpd3Δ strain was negligibly small relative to the control rrn9Δ strain (amount of Pol II bands in lane 4/amount of Pol II bands in lane 2 = 0.017). We also ascertained chromosomal rRNA gene repeat numbers per genome and found that the rrn9Δ strain used had ∼50% more than the rrn9Δ rpd3Δ strain. (N-PSW cells tend to have increased rRNA gene copy numbers even on galactose due to a slight growth advantage [see text].) This difference is small and does not affect the conclusion that the rpd3Δ mutation inhibits Pol II transcription of chromosomal rRNA genes. (B) RNA was prepared from strains NOY388 (WT), NOY1022 (uaf30Δ), NOY2015 (rpd3Δ), NOY2107 (uaf30Δ rpd3Δ), and NOY902 (rrn9Δ, PSW), which were grown in YEPD complete medium to an A600 of ∼0.5. Primer extension was carried out using 2.5 μg of RNA. (C) RNA was prepared from strain NOY388 (WT; lanes 1 and 2), NOY902 (rrn9Δ, PSW; lanes 5 and 6), and the sir2Δ derivatives of these strains (NOY1045 and NOY2108, respectively; lanes 3 and 4 and lanes 7 and 8, respectively), as described above for panel B. Primer extension was carried out in duplicate using 1.5 μg total RNA for NOY388 and NOY1045, and 5 μg total RNA for NOY902 and NOY2108. Autoradiograms are shown. The Pol I start site (+1) and Pol II start sites are indicated. In panel B, phosphorimager analysis showed the Pol II/Pol I ratios to be 0.17 for lane 2 and ∼0.05 for lane 4. We note that the rrn9Δ (PSW) strain (lane 5 in panel B and lanes 5 and 6 in panel C) was derived from a rrn9Δ (N-PSW) strain carrying a helper plasmid (similar to the one shown in lane 1 in panel A), but the helper plasmid was lost. Primer extension using RNA from cells grown in YEPG complete medium also showed the pattern identical to those shown here: that is, no RNA corresponding to band G (lanes 1 and 3 in panel A) was present.