Abstract
Cyclin D1 is overexpressed in human tumors, correlating with cellular metastasis, and is induced by activating Rho GTPases. Herein, cyclin D1-deficient mouse embryo fibroblasts (MEFs) exhibited increased adhesion and decreased motility compared with wild-type MEFs. Retroviral transduction of cyclin D1 reversed these phenotypes. Mutational analysis of cyclin D1 demonstrated that its effects on cellular adhesion and migration were independent of the pRb and p160 coactivator binding domains. Genomewide expression arrays identified a subset of genes regulated by cyclin D1, including Rho-activated kinase II (ROCKII) and thrombospondin 1 (TSP-1). cyclin D1−/− cells showed increased Rho GTP and ROCKII activity and signaling, with increased phosphorylation of LIM kinase, cofilin (Ser3), and myosin light chain 2 (Thr18/Ser19). Cyclin D1 repressed ROCKII and TSP-1 expression, and the migratory defect of cyclin D1−/− cells was reversed by ROCK inhibition or TSP-1 immunoneutralizing antibodies. cyclin E knockin to the cyclin D1−/− MEFs rescued the DNA synthesis defect of cyclin D1−/− MEFs but did not rescue either the migration defect or the abundance of ROCKII. Cyclin D1 promotes cellular motility through inhibiting ROCK signaling and repressing the metastasis suppressor TSP-1.
The human cyclin D1 gene was initially cloned as a breakpoint rearrangement within parathyroid adenoma (42). Homology of the cyclin D1 cDNA to the CLN gene in yeast suggested the PRAD1 gene encoded the regulatory subunit of a serine threonine kinase. In subsequent studies, cyclin D1 was shown to bind the retinoblastoma (pRb) protein and through physical association with the cyclin-dependent kinase 4 or 6 (cdk4 or cdk6) subunit to phosphorylate pRb. Phosphorylation of pRb by the cyclin D/cdk4 holoenzyme then alters the conformation of pRb, correlating with sequential phosphorylation by cyclin E/cdk2 and the induction of DNA synthesis. The cyclin D1 gene is overexpressed in human cancers, including breast, colon, and prostate cancer, and hematopoietic malignancies (23, 39). Targeted overexpression of cyclin D1 to the mammary gland in transgenic mice was sufficient for the induction of mammary adenocarcinoma. Cyclin D1 is overexpressed in metastatic cells (19, 30). Analysis of cyclin D1-deficient mice indicates a role for cyclin D1 in both cellular survival and DNA synthesis (3). Furthermore, cyclin D1-deficient mice are resistant to gastrointestinal tumors induced by mutation of the Apc gene (28) or tumor formation induced by either mammary-targeted Ras or ErbB2 (82). Such observations are consistent with previous studies demonstrating cyclin D1 antisense abrogates epithelial growth of ErbB2-induced tumors in vivo (34).
Mutational analysis of the human cyclin D1 cDNA has identified several distinct domains involved in binding either pRb, cdk, the p160 coactivator, and histone deacetylases (22, 23, 59). The cdk-binding domain of cyclin D1 is required for the association with cdk4 and sequential phosphorylation of pRb, which in turn, leads to the release of E2F binding proteins. The release of E2F proteins, in turn, leads to the sequential regulation of E2F-responsive genes associated with the induction of DNA synthesis. The association of cyclin D1 with the p160 coactivator SRC1 (AIB1) enhances ligand-independent ERα activity in cultured cells. Recent studies have demonstrated the regulation of several transcription factors through a cdk-independent mechanism, including MyoD, Neuro-D, the androgen receptor, CEBPβ, and peroxisome proliferator-activated receptor gamma (PPARγ) (reviewed in reference 73). The abundance of cyclin D1 is rate limiting in progression through the G1 phase of the cell cycle in fibroblasts and mammary epithelial cells. Sustained extracellular signal-regulated kinase (ERK) activation induces cyclin D1 transcription and mRNA and protein abundance, which is required for mid-G1-phase induction of cyclin D1 (2, 56, 75). Tightly coordinated interactions between the Rho GTPases facilitate cell cycle progression through regulating the expression of cyclin D1 and assembly of cyclin D/cdk complexes (12). Rac and Cdc42 induce cyclin D1 independently of ERK involving an NF-κB signaling pathway (12, 31, 79). Rho kinase suppresses Rac/Cdc42-dependent cyclin D1 induction through LIMK (56) independently of cofilin or actin polymerization. The inhibition of Rac/Cdc42 signaling maintains mid-G1-phase ERK-dependent induction of cyclin D1 (56).
The Rho family of small GTPases play an important role in the regulation of cell motility via their effects on the cellular cytoskeleton and adhesion (5, 32). Rac and its effector, PAK, induce membrane ruffles and actin rearrangements including stress fibers that control formation of lamellipodia and new focal contacts at the leading edge that drive cellular motility (54). Rho regulates assembly of stress fibers and associated focal adhesions through its downstream effectors mouse Diaphanous (mDia) and the Rho-activated kinase (ROCK) that phosphorylate cytoskeletal proteins. Major ROCK substrates regulating cellular migration include LIM kinases, which phosphorylate and regulate an actin-depolymerizing protein cofilin, and myosin light chain (MLC) kinase. Although Rho activity negatively influences cell migration by increasing stress fiber-dependent adhesions to substratum, Rho activity is also required for actomyosin contractility needed to drive cell body retraction at the rear of the cell (4). Dynamic activation and inactivation is tightly coordinated, and insufficient levels or excessive Rho GTPase activity will prevent cell migration (52, 57, 58, 71).
A variety of cytokines, chemokines, growth factors, extracellular matrix, and matrix-degrading proteins coordinate their signaling to affect migratory cues through the Rho family GTPases, and these factors are in turn regulated by Rho GTPases. Thrombospondin 1 (TSP-1), for example, is a matrix glyocoprotein that inhibits cellular metastasis and is repressed by oncogenic Ras (64). It is the first protein to be recognized as a naturally occurring inhibitor of angiogenesis (26). TSP-1 overexpression inhibits wound healing and tumorigenesis (55, 63, 64, 65). Conversely, lack of functional TSP-1 increases tissue vascularization. The abundance of TSP-1 is tightly regulated, and it is the alteration from the physiological level that seems to specifically affect migration. Thus, inhibition of TSP-1 from TSP-1-oversecreting cells reverts abnormal migration, but immunoneutralizing antibodies to TSP-1 do not affect migration of normal cells (72).
In the present study, cyclin D1−/− mouse embryo fibroblasts (MEFs) exhibit increased adherence and cellular spreading and decreased cellular motility compared with wild-type cells. Genomewide expression analysis of cyclin D1−/− MEFs transduced with a cyclin D1 expression vector that rescued the defect in adhesion and migration identified a subset of genes governing migration that were regulated by cyclin D1, including TSP-1 and ROCKII. cyclin E knockin to the cyclin D1−/− MEFs rescued the DNA synthesis defect of cyclin D1−/>− MEFs but did not rescue either the migration defect or the abundance of ROCKII. Increased ROCKII activity in cyclin D1−/− cells was corroborated by increased phosphorylation of LIM kinase, cofilin, and MLC. TSP-1 immunoneutralization or ROCK kinase inhibition rescued the migration defect of cyclin D1−/− MEFs. Thus, cyclin D1 inhibits the ROCKII and TSP-1 signaling pathway to promote cellular migration.
MATERIALS AND METHODS
Mice.
All animal experiments were performed in accordance with the guidelines for the care and use of laboratory animals at Georgetown University and Thomas Jefferson University. cyclin D1−/− mice and cyclin E knockin to the cyclin D1−/− mice were maintained as described previously (24, 28, 60). Genotyping was performed on tail DNA by PCR as described before (24, 60).
Cell culture.
Cultures of cyclin D1+/+, cyclin D1−/−, and cyclin E knockin to the cyclin D1−/− primary MEFs were prepared as described previously (74). Human kidney 293 and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing penicillin and streptomycin (100 mg of each/liter) and supplemented with 10% fetal bovine serum (FBS).
Western blotting.
Whole-cell lysates (50 μg) were separated by 10% sodium dodecyl sulfate (SDS-PAGE), and the proteins were transferred to nitrocellulose membrane for Western blotting as previously described (11). The following antibodies were used for Western blotting: rabbit Ab-3 anti-cyclin D1 antibody and mouse Ab-11 antithrombospondin (TSP) from Lab Vision/NeoMarker, Fremont, CA; an antibody against guanine nucleotide dissociation inhibitor (GDI) (35); mouse M2 anti-FLAG antibody, and antivinculin antibody from Sigma, St. Louis, MO; rabbit polyclonal antibody for Thr18/Ser19 phospho-myosin light chain 2, rabbit anti-LIMK1, and rabbit anti-phospho-LIMK (Thr508/Thr505) from Cell Signaling Technology, Beverly, MA; rabbit polyclonal antibody for Ser3 phospho-ADF/cofilin, mouse DCS-6 anti-cyclin D1 antibody, rabbit polyclonal antipaxillin antibody, anti-ROCKII, mouse anti-human cyclin E antibody (HE12), and anti-β-tubulin antibody from Santa Cruz Biotechnology, Santa Cruz, CA; and rabbit polyclonal anti-Y118 phospho-specific paxillin antibody from Biosource International, Camarillo, CA.
Retroviral production and infection.
The mouse stem cell virus (MSCV)-internal ribosome entry site (IRES)-green fluorescent protein (GFP) retrovirus vector and the ecotropic, packaging vector, pSV-ψ−E-MLV, which provides ecotropic packaging helper function, and infection methods were as described previously (45). Briefly, the coding region of mouse cyclin D1 cDNA, 3× FLAG-tagged wild-type human cyclin D1, and a series of human cyclin D1 mutant cDNAs (74) were inserted into the MSCV-IRES-GFP vector at the EcoRI site upstream of the IRES driving expression of GFP. MSCV retroviruses were prepared by transient cotransfection with helper virus into 293T cells, using calcium phosphate precipitation. The retroviral supernatants were harvested 48 h after transfection and filtered through a 0.45-μm filter. cyclin D1−/− MEFs were incubated with fresh retroviral supernatants in the presence of 8 μg/ml Polybrene for 24 h, cultured for a further 4 days, and subjected to fluorescence-activated cell sorting (FACS) (FACStar Plus; BD Biosciences, San Jose, CA) to select for cells expressing GFP. GFP+ cells were used for subsequent analysis.
Fluorescent and phase-contrast light microscopy.
GFP+ cells were examined in six-well plates. Fluorescent microscopy and phase-contrast imaging were carried out using the ×10 objective of an Olympus IX-70 laser-scanning confocal microscope.
Cell cycle analysis, phalloidin staining, and F-actin quantitation.
DNA synthesis of MEFs was determined by propidium iodide (PI) staining and FACS analysis as described previously (1). Phalloidin staining was conducted as previously described (45). F-actin quantitation was carried out by FACS analysis (50).
Immunofluorescence.
cyclin D1+/+, cyclin D1−/−, and cyclin D1−/− MEFs infected with MSCV-cyclin D1-IRES-GFP or GFP vector control grown in four-well chamber slides were fixed with cold methanol for 20 min at 4°C. The slides were air dried at room temperature. The primary antibodies used were mouse monoclonal antipaxillin (clone 5H11; Upstate) (1/100) and rabbit polyclonal anti-phospho-paxillin (pY118) (Biosource) (1/100). The secondary antibodies used were Alexa Fluor 488-conjugated F(ab′)2 fragment of goat anti-mouse immunoglobulin G (IgG) (Molecular Probes, Inc.) (1/250) and rhodamine red X-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Lab.) (1/50). The samples were visualized on an Olympus IX70 laser-scanning confocal microscope with a ×60 PlanApo oil immersion objective and using Olympus Fluoview FV-300 software. The images were processed with MetaMorph (Molecular Devices).
SEM.
For scanning electron microscopy (SEM), cells were plated on fibronectin-coated glass coverslips and grown to approximately 80% confluence. The cells were fixed as described previously (45) to prevent agonal membrane artifacts. Dehydrated cells were critical point dried using liquid carbon dioxide in a Samdri 790 critical point drier (Tousimis Research, Rockville, MD), sputter-coated with gold-palladium in a Vacuum Desk-1 sputter coater (Denton, Cherry Hill, NJ), mounted, and viewed in a AMRAY 1820D scanning electron microscope by using an accelerated voltage of 20 kV for electron microscopy.
IRM.
Interference reflection microscopy (IRM) was performed as described as previously (45). Briefly, IRM was performed using a ×60 objective of an Olympus IX-70 laser-scanning confocal microscope in reflectance mode with polarization filters at either 488 or 568 nm. Direct adherence or apposition of the cell to the substrate is imaged as black (destructive interference), and greater distance is viewed as white (direct reflection or constructive interference). Image analysis was performed on the images based on the assumption that more adherent cells would have more dark pixels per unit area of spread than less-adherent cells. NIH ImageJ software was used to manually trace the boundaries of individual cells, and a grayscale histogram was derived for each cell. The threshold was set based on the shape of the histogram (15). Values below the threshold corresponded to regions that appeared black. A ratio was derived for each cell (percentage close apposition) that consisted of the sum of pixels below the threshold divided by the total number of pixels within the boundaries of the cells.
Adhesion assay.
Adhesion assays were conducted as previously described (45). Briefly, cells (2 × 104) were seeded in serum-free medium in a 96-well plate coated with 10 μg/ml fibronectin (Sigma, St. Louis, MO). After 30 min, plates were washed twice with phosphate-buffered saline (PBS), fixed, and stained with crystal violet. Adherent cells were represented as A550.
Spreading assay.
Spreading assays were conducted as described previously (47). Briefly, cells were plated on 60-mm plastic tissue culture dishes in DMEM containing 10% FBS for the indicated time points. Dark cells were considered to be spread, and bright cells were considered to be unspread. Pictures of three independent fields were taken under the ×10 objective. Experiments were done in triplicate and repeated at least three times.
Migration assay.
Transwell migration assays were performed as described before (38). Briefly, GFP-positive cells were seeded on 8-μm-pore-size Transwell filter insert (Costar) coated with 10 μg/ml fibronectin (Sigma, St. Louis, MO). After 16 h of incubation at 37°C and 5% CO2, cells adherent to the upper surface of the filter were removed using a cotton applicator. Cells were fixed with 3.7% formaldehyde and stained with crystal violet, and the numbers of cells on the bottom were counted. Data are from three experiments done in triplicate (mean ± standard error).
Wound healing.
Cells were grown to confluence on 12-well plates, and the monolayers were wounded with a P10 micropipette tip (45). DMEM with 10% FBS and HEPES was changed immediately after scoring. The wound-healing videos were taken at 20-min intervals (30 min in p16INK4a peptide experiments) using a Nikon Eclipse TE-300 inverted microscope system. The cell movement velocity was determined by tracing single cells at different time points using MetaMorph software (Molecular Devices Corporation, Downington, PA).
p16 peptides.
Peptides corresponding to amino acids 84 to 103 of human p16INK4a protein with a C-terminal sequence of 16 amino acids encoding the Antennapedia homeodomain (Penetratin) (20, 21, 27) were synthesized (Bio-synthesis, Inc. Lewisville, TX). Peptide 20 (DAAREGFLATLVVLHRAGARRQIKIWFQNRRMKWKK) with the D92A substitution has a lower 50% inhibitory concentration (IC50) to inhibit cdk4-cyclin D1 phosphorylation of a glutathione S-transferase (GST)-Rb protein in vitro and to arrest cell cycle progression in G1 than the corresponding peptide containing the wild-type sequence (20, 21). Peptide 21 (DAAREGFLDTLAALHRAGARRQIKIWFQNRRMKWKK) carrying VV95 and 96AA mutations, has an increased IC50 in vitro and has lost ∼60% of the cell cycle inhibitory capacity (20, 21). These peptides were added to the cell culture medium at a concentration of 20 μM.
Time-lapse video.
For time-lapse observation of cell movement, cells on 12-well plates were maintained in DMEM with 10% fetal calf serum (FCS) and HEPES. Cells were placed in a temperature and CO2 control incubator to maintain the temperature at 37°C and CO2 at 5%. The cell movement videos were taken at 5-min intervals by using a Nikon Eclipse TE-300 inverted microscope system. The cell movement velocity was determined by tracing the single cells at different time points using MetaMorph software. To observe the effect of ROCK inhibition, cells were treated with 10 μM Y27632 (Calbiochem) for 30 min before time-lapse recording. For assays with neutralizing antibody, the A4.1 anti-TSP-1 monoclonal antibody (Neomarkers) was added at 10 μg/ml as previously described (72).
Microarray analysis.
Total RNA was isolated from retrovirus vector-infected cyclin D1−/− MEFs (infected with either MSCV-cyclin D1-IRES-GFP or MSCV-IRES-GFP control vector) using Trizol and used to probe Affymetrix MU74v2 arrays (Affymetrix, Santa Clara, CA). RNA quality was determined by gel electrophoresis. Probe synthesis and hybridization were performed according to the manufacturer's manual (see the eukaryotic target preparation section at http://www.affymetrix.com/support). Three arrays were used for each condition. Analysis of the arrays was performed using the statistical package R statistics package (14) and the limma library (62) of the Bioconductor software package. Arrays were normalized using robust multiarray analysis (RMA), and the genes were ranked using the log odds ratios for differential expression (9, 53). Briefly, a linear model was constructed using a factorial design and differentially expressed genes were obtained from cyclin D1−/− plus cyclin D1 or a cyclin D1−/− vector control. Finally, the top-ranked genes that are differentially expressed in a cyclin D1-dependent manner were determined based on their log odds ratio. These genes were then clustered using hierarchical clustering with “complete” agglomeration, and each cluster was further analyzed based upon the known function of the genes contained in the cluster. All six arrays were analyzed and show good separation based upon their cyclin D1 expression levels (8, 62).
Luciferase assays.
A luciferase reporter plasmid containing bp −2800 to +48 of the murine thrombospondin 1 (mTSP-1) promoter was kindly provided by Paul Bornstein (University of Washington, Seattle) (7). HEK 293 cells were used in mTSP-1 reporter assays. The cells were transiently transfected with increasing amounts of wild-type or KE mutant cyclin D1 (in a cytomegalovirus 10 [CMV10] vector) in combination with the reporter using Superfect reagent (QIAGEN, Valencia, CA), according to the manufacturer's instructions. Forty-eight hours posttransfection, luciferase assays were performed at room temperature using an Autolumat LB 953 (EG&G Berthold) as previously described (74). A CMV-β-galactosidase plasmid was cotransfected as a control for transfection efficiency. Luciferase activities were measured and normalized by the empty vector CMV10 control.
Semiquantitative RT-PCR.
Total cellular RNA was prepared from cyclin D1+/+ and cyclin D1−/− P-3 MEFs. Semiquantitative reverse transcription-PCR (RT-PCR) was performed as previously described (72) using the following primers: forward primer 5′ GGGCTAGAGAAACCCCCCAC 3′ and reverse primer 5′ CCAAAGGGAGAAAGTCC 3′ to amplify murine TSP-1 and forward primer 5′ TGTTACCAACTGGGACGACA 3′ and reverse primer 5′ AAGGAAGGCTGGAAAAGAGC 3′ to amplify murine β-actin.
Rho pulldown assay.
The amount of activated Rho present in cyclin D1−/− MEFs infected with wild-type cyclin D1 and vector was determined by using the Rho activation assay kit (Upstate Cell Signaling Solutions, Lake Placid, NY), according to the manufacturer's instruction. The amounts of bound (activated) and total Rho (RhoA, -B, and -C) were measured by immunoblotting.
ROCKII kinase assay.
For ROCKII kinase activity assays, cell lysates (500 μg protein in NP40 lysis buffer: 1% NP40, 150 mM NaCl, 50 mM Tris, pH 7.4) were incubated with 10 μl of polyclonal anti-ROCKII (H-85; Santa Cruz Biotechnology) and 50 μl protein A agarose (Roche) overnight at 4°C. The immunocomplexes were washed and subjected to kinase reaction in 20 μl of kinase buffer (50 mM Tris, pH 7.4, 10 mM MgCl2, 3 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA) in the presence of 10 μCi of [γ-32P]ATP and 20 μg of myelin basic protein (MBP) (Sigma). Reaction mixtures were incubated for 20 min at 37°C, and reactions were terminated by the addition of 4 μl of 6× SDS sample buffer. Samples were resolved by 15% SDS-polyacrylamide gel electrophoresis, and the gel was dried and subjected to autoradiography.
Online supplemental material.
An online supplement includes the wound-healing videos (http://www.kimmelcancercenter.org/pestell/papers/zl/video/) of cyclin D1+/+ (see Fig. 5Avideo1 at the URL mentioned above), cyclin D1−/− MEFs (Fig. 5Avideo2), cyclin D1−/− MEFs infected with wild type (Fig. 5Bvideo1) or mutant GH (Fig. 5Bvideo2), LLAA (Fig. 5Bvideo3), KE (Fig. 5Bvideo5) cyclin D1 or vector control (Fig. 5Bvideo4), and cyclin D1+/+ MEFs treated with p16INK4a peptide 20 (Fig. 5Dvideo1) or p16INK4a peptide 21 (Fig. 5Dvideo3) or without peptide treatment control (Fig. 5Dvideo2). Microarray data of cyclin D1−/− plus cyclin D1 and vector control were also included in the online supplemental material (http://www.kimmelcancercenter.org/pestell/papers/zl/D1vsGFP.xls). Supplements 1 and 2 may also be found online (http://www.kimmelcancercenter.org/pestell/papers/zl/Supplement1.pdf and http://www.kimmelcancercenter.org/pestell/papers/zl/Supplement2.pdf).
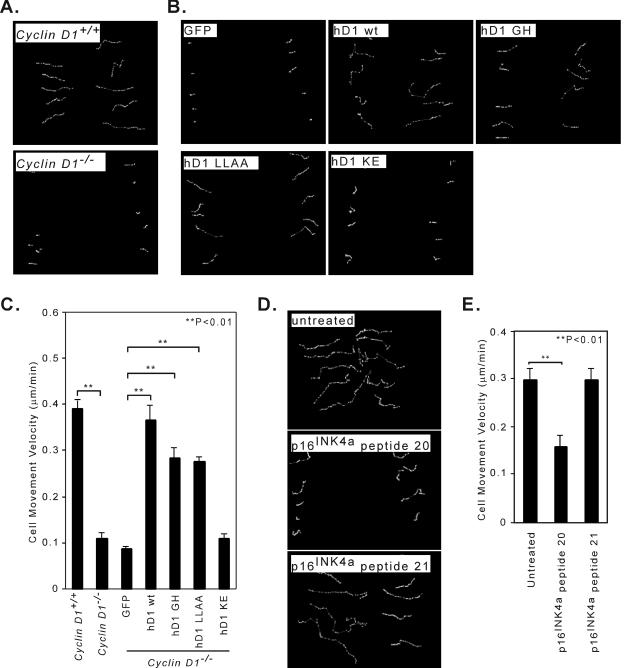
FIG. 5.
Cyclin D1 regulation of wound closure requires the cyclin D1 cdk-binding domain. (A) Cyclin D1-deficient MEFs display a decreased motility as detected in the wound-healing assay. cyclin D1+/+ and cyclin D1−/− MEFs were grown to confluence on 12-well plates, and the monolayers were wounded with a P10 pipette tip. Video images were recorded at 20-min intervals using a Nikon Eclipse TE-300 inverted microscope system. (B) cyclin D1−/− MEFs transduced with either MSCV-GFP or the cyclin D1 wild type or mutants were assessed for wound closure. The velocity of cell movement was determined by tracing the movement of single cells with time using MetaMorph software. (C) Data are presented as the mean ± standard error of at least 6 individual cells of the same genotype. ** indicates the significant difference between cyclin D1+/+ and cyclin D1−/− MEFs, and the significant difference between wild-type (wt) or mutant cyclin D1-infected cells and control GFP vector-infected cells. (D) cyclin D1+/+ MEFs treated with p16INK4a peptide 20 or p16INK4a peptide 21 or without peptide treatment were assessed for wound closure. cyclin D1+/+ MEFs were grown to confluence on 12-well plates, and the monolayers were wounded with a P10 pipette tip. The wound-healing video images were recorded at 30-min intervals. The cell movement velocity was determined by tracing the single cells at different time points using MetaMorph software. (E) The data for cell movement velocity are shown as the mean ± standard error of at least 6 individual cells of the same genotype. **, significant difference between peptide-treated cells and untreated cells.
RESULTS
MEF morphology is changed by cyclin D1 deficiency, and the cyclin D1 KE mutant failed to rescue this defect.
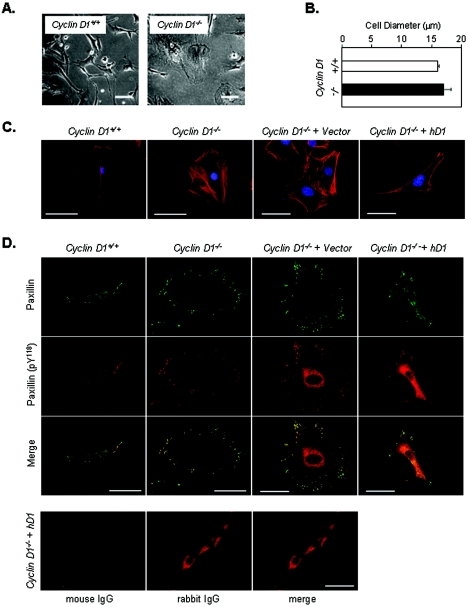
By phase-contrast microscopy, cyclin D1−/− MEFs were more spread than wild-type MEFs (Fig. 1A), with no significant difference in cell diameter in suspension, assessed using a Multisizer 3 Coulter counter (Fig. 1B). The flatter, rounder, less elongated morphology of the cyclin D1−/− MEFs was accompanied by an increased F-actin with a cortical distribution (Fig. 1C) of F-actin and a reduction in cytoplasmic microfilament bundling. The MSCV-GFP retrovirus vector-infected cyclin D1−/− MEFs with high efficiency without affecting cellular morphology, whereas the reintroduction of human or murine cyclin D1 restored the elongated morphology and rescued the loss of cytoplasmic stress fibers (Fig. 1C). F-actin abundance by FACS analysis (data not shown) was not significantly different between cyclin D1+/+ and cyclin D1−/− MEFs or between cyclin D1−/− MEFs infected with cyclin D1 and GFP vector control, suggesting altered F-actin distribution rather than altered gene expression contributes to the less elongated phenotype in cyclin D1−/− MEFs. In wild-type MEFs, infrequent costaining for paxillin and tyrosine-phosphorylated paxillin (Y118) was identified. In contrast, sites of tyrosine-phosphorylated paxillin were increased in number and centripetally distributed around the circumference of cyclin D1-deficient cells (Fig. 1D). Reintroduction of cyclin D1 reduced the number and reversed the circumferential distribution of focal contacts. Control IgG showed only background staining (not shown). Overexposed images of IgG control staining showed no costaining sites (Fig. 1D).
FIG. 1.
Cyclin D1-deficient cells have increased stress fiber formation. (A) cyclin D1+/+ and cyclin D1−/− MEFs were plated on six-well plates and examined by phase-contrast microscopy (×10). Scale bar, 50 μm. (B) Cellular diameter assessed by Multisizer 3 Coulter counter. (C) Rhodamine-phalloidin staining of cyclin D1+/+ or cyclin D1−/− MEFs and cyclin D1−/− MEFs transduced with control vector (GFP) or human cyclin D1 (hD1wt). Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI). Scale bar, 50 μm. (D) Immunofluorescence analysis of cyclin D1+/+ or cyclin D1−/− MEFs and cyclin D1−/− MEFs transduced with vectors encoding human cyclin D1 (hD1wt) or vector control with antibodies for pY118-paxillin (red) and paxillin (green). Yellow dots indicate their colocalization. Typical examples of representative cells are shown. Note the centripetal distribution of tyrosine-phosphorylated paxillin in cyclin D1-deficient cells. Scale bar, 50 μm.
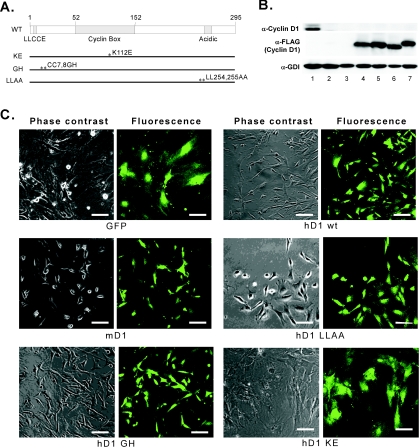
Murine cyclin D1 (data not shown) and wild-type or mutant human cyclin D1 proteins were expressed in cyclin D1−/− MEFs (Fig. 2A and B). Expression of human or murine cyclin D1 conferred wild-type cellular morphology to cyclin D1−/− MEFs (Fig. 2C). Expression of either the cyclin D1 GH mutant, which is defective in pRb binding, or the cyclin D1 LLAA mutant, which fails to bind the p160 coactivator SRC1 (AIB1), rescued the cell morphology phenotype. However, this phenotype was not rescued by the cdk4-binding-defective mutant of cyclin D1 (KE mutant).
FIG. 2.
The cyclin D1 domains required for rescue of cellular morphology. (A) Schematic representation of wild-type (WT) and mutant human cyclin D1 expression vector structure. (B) Whole-cell lysates were examined by SDS-PAGE and Western blotting for endogenous cyclin D1, FLAG-tagged cyclin D1, and GDI (loading control) expression. Lane 1, cyclin D1+/+ MEFs; lane 2, cyclin D1−/− MEFs; lane 3, cyclin D1−/− MEFs infected with MSCV-GFP vector control; lane 4, cyclin D1−/− MEFs infected with human cyclin D1 wild type; lane 5, cyclin D1−/− MEFs infected with human cyclin D1 KE mutant; lane 6, cyclin D1−/− MEFs infected with human cyclin D1 GH mutant; lane 7, cyclin D1−/− MEFs infected with human cyclin D1 LLAA mutant. (C) Phase-contrast and fluorescence microscopy images (×10) were taken from cyclin D1−/− MEFs infected with either MSCV-GFP vector or MSCV-wild-type (wt) or mutant cyclin D1 expression vectors, as indicated. mD1, mouse cyclin D1. Scale bar, 50 μm.
The cyclin D1 cdk-binding domain is required for reducing cellular spreading and inducing DNA synthesis.
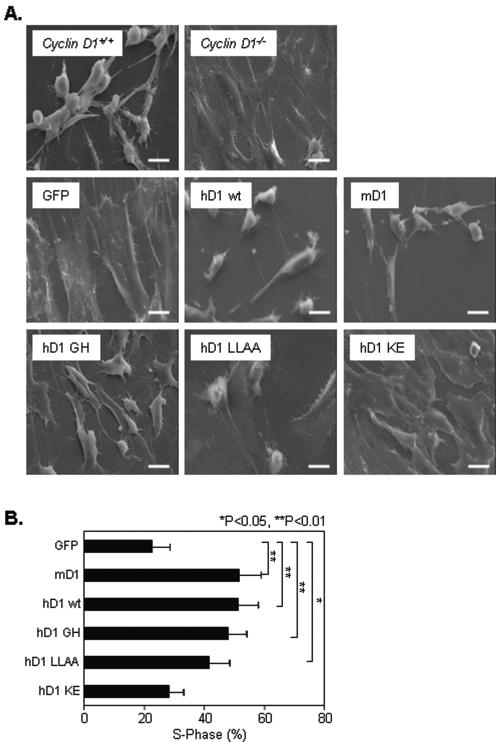
By scanning electron microscopy (Fig. 3A), cyclin D1−/− MEFs were more spread than cyclin D1+/+ MEFs. Expression of either murine or human cyclin D1 in cyclin D1−/− MEFs rescued the wild-type MEF phenotype. The pRb-binding-defective mutant (GH mutant) and SRC1-binding-defective mutant (LLAA mutant) were both capable of rescuing this phenotype, whereas the cdk-binding-defective mutant (KE mutant) was again unable to rescue (Fig. 3A). cyclin D1−/− MEFs and 3T3 cells exhibited a reduced proportion of cells in S phase of the cell cycle (3). Expression of cyclin D1 wild-type or the pRb-binding-defective or SRC1-binding-defective mutants increased the proportion of cells in S phase from 22% to up to 51% (Fig. 3B) (P ≤ 0.02). Mutation of the cdk4-binding site in cyclin D1 abrogated this rescue of DNA synthesis. All cyclin D1−/− MEFs transduced with the MSCV-cyclin D1-IRES-GFP vector reverted to the fibroblastoid morphology, yet only 50% of the cells were in the DNA synthetic phase.
FIG. 3.
The cdk-binding domain of cyclin D1 regulates cellular spreading. (A) cyclin D1+/+, cyclin D1−/−, and cyclin D1−/− MEFs infected with either MSCV-GFP vector or wild-type or mutant mouse (mD1) or human (hD1) cyclin D1 were plated on fibronectin (10 μg/ml)-coated coverslips and examined by scanning electron microscopy (×1,000). Scale bar, 20 μm. (B) DNA synthesis and cell cycle analysis of MEFs were performed by propidium iodide staining and FACS analysis. Cells were either infected with MSCV-GFP vector control or viruses encoding murine or human cyclin D1 wild type (wt) or mutants, as indicated. * and ** indicate significant difference between wild-type or mutant cyclin D1-infected cells and MSCV-GFP control cells.
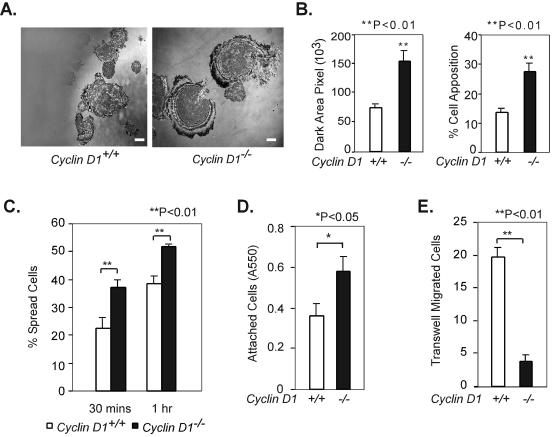
The close apposition of cell with substratum is inhibited by cyclin D1 through the cdk-binding domain.
IRM has been used to measure the appositional proximity of cells with the substratum (45). Cell areas with close contact with the substratum, including focal adhesions, are displayed as dark structures, whereas less closely apposed regions exhibit various degrees of gray and white. cyclin D1−/− MEFs showed increased dark area pixels, and the dark area pixels as a percentage of the total area pixels was also enhanced compared to that in wild-type cells (Fig. 4A and B) (75,080 ± 4,840 versus 154,280 ± 17,840, 14% ± 0.8% versus 27% ± 3%) (P = 2.7 × 10−5 and P = 3.5 × 10−5, respectively), indicating that cyclin D1−/− MEFs show enhanced cell-substratum apposition. Expression of mouse or human cyclin D1 or the pRb-binding defective (GH) or the SRC1-binding defective (LLAA) mutants of cyclin D1 in cyclin D1−/− MEFs significantly reduced the dark area of apposition (see supplement 1A at http://www.kimmelcancercenter.org/pestell/papers/zl/Supplement1.pdf) (P ≤ 0.008). In contrast, expression of the cdk-binding defective (KE) mutant, failed to reduce the closely apposed area detected by IRM (P = 0.12 and P = 0.14, respectively) (see supplement 1B at http://www.kimmelcancercenter.org/pestell/papers/zl/Supplement1.pdf).
FIG. 4.
Cyclin D1 regulates cellular adhesion and migration. (A) cyclin D1−/− MEFs display increased apposition detected by interference reflection microscopy (60×). Scale bar, 20 μm. (B) Quantitation of dark area pixel and % cell apposition (the ratio of dark area pixel to the total area pixel) of cyclin D1+/+ and cyclin D1−/− MEFs by ImageJ software (http://rsb.info.nih.gov/ij/). (C) cyclin D1−/− MEFs spread more rapidly than cyclin D1+/+ MEFs. cyclin D1+/+ and cyclin D1−/− MEFs were assessed by enumerating dark and bright cells under phase contrast. Dark cells were considered to be spread, and bright cells were considered to be unspread. The mean ± standard error of the number of spread cells is shown at the time points 30 min and 1 h after plating. (D) cyclin D1−/− MEFs are more adherent than wild-type MEFs. A total of 20,000 wild-type and cyclin D1−/− MEFs were seeded in a 96-well plate coated with 10 μg/ml fibronectin. After 30 min, plates were washed two times with PBS, fixed, and stained with crystal violet, and the absorbance was read at 550 nm (A550). (E) cyclin D1−/− MEFs migrate more slowly than wild-type MEFs. A total of 40,000 cells were seeded in a fibronectin-coated Transwell. Cellular migration was determined by the cell number at the bottom of the Transwell plate as described in Materials and Methods.
cyclin D1−/− MEFs spread significantly more quickly than wild-type cells at 30 min and 1 h, as judged by the ratio of dark to bright cells under phase-contrast microscopy (P = 0.009 and P = 0.003, respectively) (Fig. 4C). Introduction of either the murine or human cyclin D1 cDNA into cyclin D1−/− MEFs by retroviral infection reduced the percentage of spread cells from 58% to ∼30% at 30 min after plating (P = 0.0007 and P = 0.0004, respectively) (see supplement 2 at http://www.kimmelcancercenter.org/pestell/papers/zl/Supplement2.pdf). Both the pRb-binding-defective mutant (GH mutant) and the SRC1-binding-defective mutant (LLAA mutant) decreased the percentage of spread cells at 30 min (P = 0.0032) (see supplement 2 at the URL above). However, the cdk-binding-defective mutant (KE mutant) failed to reduce the percentage of spread cells (see supplement 2 at the URL above). Cyclin D1-deficient MEFs were more adherent than wild-type cells on fibronectin (P = 0.03) (Fig. 4D). Exogenous mouse and human cyclin D1 wild-type, GH mutant, or LLAA cyclin D1 mutants decreased adherence on fibronectin (P = 8.7 × 10−10, P = 1 × 10−6, P = 9.1 × 10−6, P = 4.4 × 10−6) (see supplement 2 at the URL above), while the cdk-binding-defective mutant (KE mutant) failed to reduce adhesion (see supplement 2 at the URL above).
p16INK4a inhibits and cyclin D1 enhances the velocity of cellular migration.
cyclin D1−/− MEFs migrated significantly less than wild-type cells on fibronectin-coated Transwell plates (P = 0.0008) (Fig. 4E). Retroviral introduction of the murine or human cyclin D1 cDNA into cyclin D1−/− MEFs increased the cell migration rate significantly (P = 0.0006 and P = 0.001, respectively) (see supplement 2C at the URL above). The pRb-binding-defective cyclin D1 mutant (GH mutant) and the SRC1-binding-defective mutant (LLAA mutant) both increased cell migration (P = 0.016 and P = 0.0005, respectively) (see supplement 2C at the URL above), which was not enhanced by the cdk-binding-defective mutant (KE mutant).
By overnight time-lapse videomicroscopy, the single-cell motility of cyclin D1−/− MEFs was significantly slower than the motility of wild-type cells (P = 0.0006) (Fig. 5A; see Fig. 5Avideo1 and -2 at http://www.kimmelcancercenter.org/pestell/papers/zl/video/). Reintroduction of the cyclin D1 wild type or the cyclin D1 mutants GH or LLAA increased the velocity of cell movement compared with that in vector control cells (P = 0.0048, P = 0.0009, and P = 0.0002, respectively) (Fig. 5B; see Fig. 5Bvideo1 to -4 at http://www.kimmelcancercenter.org/pestell/papers/zl/video/), whereas there was no significant effect of the cdk-binding-defective mutant (KE mutant) (P = 0.125) (Fig. 5B; see Fig. 5Bvideo5 at http://www.kimmelcancercenter.org/pestell/papers/zl/video/). Thus, cyclin D1 enhances the velocity of cell motility.
A synthetic peptide that spans the two α-helixes of the third ankyrin repeat of p16INK4a, interacts with the cdk, inhibiting cyclin D1-kinase activity. The derivative of this peptide carrying a D92A substitution (peptide 20) (Fig. 5D) has a lower IC50 compared with the wild-type sequence (20). The p16INK4a peptide 20 linked to the Antennapedia carrier sequence inhibited wound closure compared with the no-peptide-treatment control (P = 0.0016) (Fig. 5D; see Fig. 5Dvideo1 and -2 at http://www.kimmelcancercenter.org/pestell/papers/zl/video/). The p16INK4a peptide 21, which carries alanine substitutions of two valine residues corresponding to positions 95 and 96 of the p16INK4a that dramatically increases its IC50 in vitro (20), had reduced ability to inhibit MEF migration (P = 0.98) (Fig. 5E; see Fig. 5Dvideo3 at http://www.kimmelcancercenter.org/pestell/papers/zl/video/). This finding is consistent with a model in which cdk binding regulates MEF cellular migration by cyclin D1.
Cyclin D1 regulates a molecular genetic cluster governing cellular adhesion and migration.
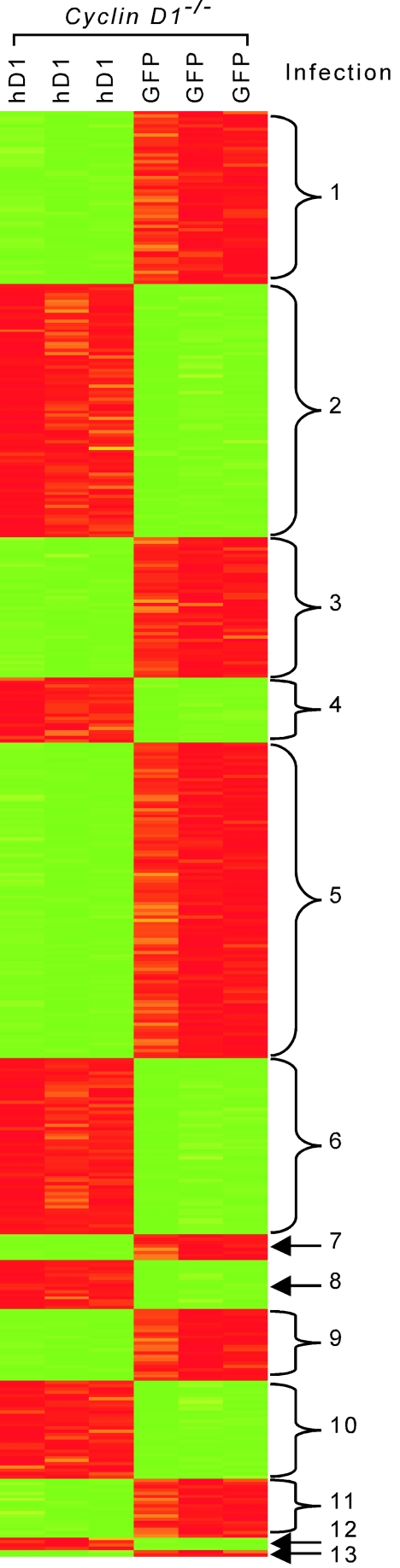
We conducted a comprehensive genomewide interrogation to identify gene targets regulated by cyclin D1 that may play a role in cell adhesion and migration. cyclin D1−/− MEFs were infected with either wild-type cyclin D1 or the empty GFP vector as control. Analyses were conducted in triplicate, and the subsets of genes differentially regulated by cyclin D1 were identified (Fig. 6 and Table 1; see also http://www.kimmelcancercenter.org/pestell/papers/zl/D1vsGFP.xls). Differential expression of genes previously identified as regulating cellular migration and adhesion included those coding for TSP-1 and ROCKII. Given the central role of ROCKII and TSP-1 in regulating cellular migration, we considered the possibility that cyclin D1 may regulate the abundance and activity of these two key proteins.
FIG. 6.
Summary of genes regulated by cyclin D1 expression. Total RNA isolated from retrovirus-infected cyclin D1−/− MEFs (MSCV-cyclin D1-IRES-GFP or MSCV-IRES-GFP vector control) was used to probe the Affymetrix murine genome U74v2 set. The probes that correspond to a log odds ratio greater than 4 or a probability of differential expression greater than 0.95 were then hierarchically clustered. Data from each probe are in the rows, and each experiment is shown as a column. Red and green denote increased and decreased expression levels respectively, with the intensity reflecting the magnitude of change. Thirteen groups of genes are shown in the figure. The genes in each group are shown online at http://www.kimmelcancercenter.org/pestell/papers/zl/GenesFigure8.xls.
TABLE 1.
Adhesion- and migration-related genes regulated by cyclin D1a
| Protein designation | Name | GenBank accession no. | Fold change in cyclin D1 vs GFP vector | Bb | Probability of differential expression (%) |
|---|---|---|---|---|---|
| Ctgf | Connective tissue growth factor | NM_010217 | −4.53 | 2.11E + 01 | 100.00 |
| Ptgs2 | Prostaglandin-endoperoxide synthase 2 (cyclooxygenase 2) | NM_011198 | −4.35 | 1.62E + 01 | 100.00 |
| Tnc | Tenascin C (hexabrachion) | NM_011607 | −3.86 | 1.99E + 01 | 100.00 |
| Selp | Selectin P (granule membrane protein 140 kDa, antigen CD62) | NM_011347 | −3.25 | 1.64E + 01 | 100.00 |
| Col8a1 | Collagen, type VIII, alpha 1 | NM_007739 | −2.57 | 1.65E + 01 | 100.00 |
| Fez1 | Fasciculation and elongation protein zeta 1 (zygin I) | NM_183171 | −2.35 | 1.38E + 01 | 99.99 |
| Rhob | Ras homolog gene family, member B | NM_007483 | −2.2 | 1.35E + 01 | 99.99 |
| TSP-1 | Thrombospondin 1 | NM_011580 | −2.11 | 6.12E + 00 | 98.58 |
| Anxa1 | Annexin A1 | NM_010730 | −1.9 | 5.53E + 00 | 97.88 |
| F11r | F11 receptor | NM_172647 | −1.85 | 1.05E + 01 | 99.93 |
| Cdh2 | Cadherin 2, type 1, N-cadherin (neuronal) | NM_007664 | −1.8 | 7.73E + 00 | 99.53 |
| Pkp2 | Plakophilin 2 | NM_026163 | −1.67 | 6.15E + 00 | 98.61 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | NM_011333 | −1.65 | 8.16E + 00 | 99.65 |
| Mcam | Melanoma cell adhesion molecule | NM_023061 | −1.59 | 5.80E + 00 | 98.24 |
| ROCKII | Rho-associated coiled-coil-forming kinase 2 | NM_009072 | −1.57 | 5.85E + 00 | 98.30 |
| Tgfb1i1 | Transforming growth factor-β1-induced transcript 1 | NM_009365 | −1.56 | 9.63E + 00 | 99.87 |
| Fbln2 | Fibulin 2 | NM_007992 | −1.53 | 8.76E + 00 | 99.77 |
| Tln1 | Talin 1 | NM_011602 | −1.5 | 4.86E + 00 | 96.67 |
| Map2k1 | Mitogen-activated protein kinase kinase 1 | NM_008927 | −1.48 | 5.94E + 00 | 98.4 |
| Jup | Junction plakoglobin | NM_010593 | −1.43 | 4.54E + 00 | 95.88 |
| Fxyd5 | FXYD domain containing ion transport regulator 5 | NM_008761 | −1.37 | 4.44E + 00 | 95.6 |
| Fbln1 | Fibulin 1 | AK083573 | 1.36 | 4.28E + 00 | 95.1 |
| Serpine 2 | Serine (or cysteine) proteinase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 | NM_009255 | 1.59 | 6.23E + 00 | 98.69 |
| Islr | Immunoglobulin superfamily containing leucine-rich repeat | NM_012043 | 1.61 | 8.31E + 00 | 99.69 |
| Gas6 | Growth arrest-specific 6 | NM_019521 | 1.63 | 8.36E + 00 | 99.70 |
| Ccl5 | Chemokine (C-C motif) ligand 5 | NM_013653 | 1.69 | 1.04E + 01 | 99.93 |
| Col6a1 | Collagen, type VI, alpha 1 | NM_009933 | 1.7 | 9.55E + 00 | 99.87 |
| Cx3cl1 | Chemokine (C-X3-C motif) ligand 1 | NM_009142 | 1.98 | 1.29E + 01 | 99.99 |
| Col6a2 | Collagen, type VI, alpha 2 | NM_146007 | 2.2 | 1.42E + 01 | 99.99 |
| Tpbg | Trophoblast glycoprotein | NM_011627 | 2.22 | 1.28E + 01 | 99.99 |
| Vegfc | Vascular endothelial growth factor C | NM_009506 | 2.81 | 1.15E + 01 | 99.97 |
| Tgfbi | Transforming growth factor, beta induced, 68 kDa | NM_009369 | 3.01 | 1.40E + 01 | 99.99 |
Shown are genes differentially regulated by cyclin D1, which have adhesion- and/or migration-related function.
B is the log odds ratio of the gene being differentially expressed.
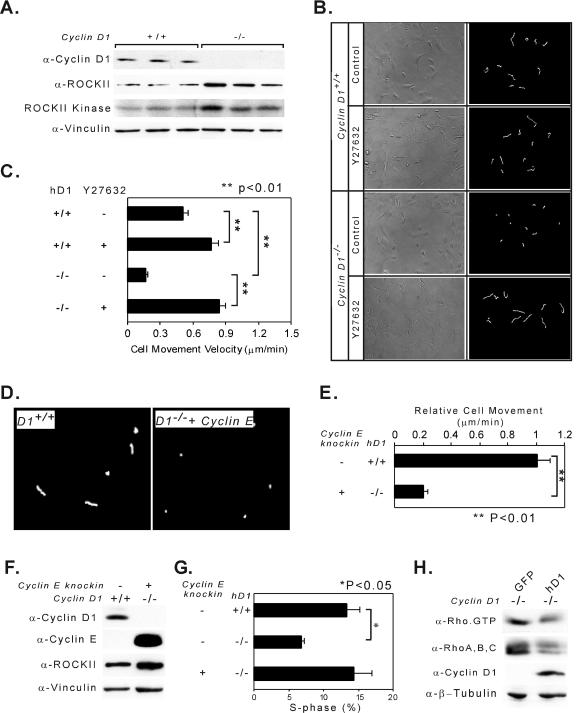
The increase in actin stress fiber and focal contact formation in cyclin D1−/− MEFs is consistent with prior studies of cells with increased ROCK activity (13, 70). Consistent with the microarray analysis, ROCKII levels were increased in cyclin D1−/− MEFs compared to sibling controls. ROCKII kinase activity was increased in cyclin D1−/− cells (Fig. 7A). Addition of the ROCK kinase inhibitor Y-27632 increased cyclin D1−/− cellular migration and movement velocity ∼6-fold (Fig. 7B and C). To determine whether the cyclin E could rescue the migration defect of cyclin D1 deficiency, we examined MEFs derived from mice in which the cyclin D1 gene coding sequence was replaced with the human cyclin E gene cDNA (cyclin E knockin to the cyclin D1−/− mice) (24). The cell movement velocity of the cyclin E knockin MEFs was reduced to approximately 20% of that of wild-type littermate controls (Fig. 7D and E). Cyclin E protein was well expressed in the cyclin E knockin MEFs (Fig. 7F); however, as with the cyclin D1−/− MEFs, ROCKII abundance remained increased. The cyclin E knockin was capable of rescuing the defect in DNA synthesis of the cyclin D1−/− cells (Fig. 7G), as previously described (24). The reduction in ROCK II kinase activity in the cyclin D1+/+ cells compared with cyclin D1−/− cells suggested cyclin D1 inhibited Rho activity. To determine whether cyclin D1 was sufficient to inhibit Rho GTPase activity, cyclin D1−/− cells were transduced with a cyclin D1-expressing retrovirus. Consistent with the relative reduction in ROCKII kinase in the cyclin D1-expressing cells, Rho-GTP activity and total Rho abundance were decreased upon cyclin D1 transduction of cyclin D1−/− cells compared to vector control-transduced cells (Fig. 7H).
FIG. 7.
Cyclin D1 promotes migration through inhibition of ROCK kinase. (A) Western blot analysis and ROCKII kinase assays in three pairs of cyclin D1+/+ and cyclin D1−/− MEFs. Each lane represents a different animal. Vinculin was used as a loading control. α-, anti-. (B) Single-cell analysis of cell movement velocity taken at 5-min intervals from cyclin D1+/+ or cyclin D1−/− MEFs treated with either vehicle or the ROCK kinase inhibitor Y27632. (C) Cellular movement velocity, shown as mean ± standard error for n ≥ 20 cells (**, P < 0.01) assessed at 5-min intervals for 2 h. (D and E) Single-cell analysis of cell movement velocity was taken at 5-min intervals from cyclin D1+/+ or cyclin E knockin to cyclin D1−/− MEFs. Cellular movement velocity is shown as the mean ± standard error for n ≥ 10 cells (**, P < 0.01). (F) Western blot analysis for ROCKII protein expression in cyclin D1+/+ and cyclin E knockin to cyclin D1−/− MEFs. Vinculin was used as a loading control. (G) DNA synthesis and cell cycle analysis of cyclin D1+/+, cyclin D1−/− MEFs, and cyclin D1−/− with human cyclin E knockin MEFs were performed by propidium iodide staining and FACS analysis. An asterisk indicates the significant difference between cyclin D1+/+ and cyclin D1−/− MEFs. (H) Rho protein and activated Rho decreased by cyclin D1. Rho-GTP pull-down assay and Western blot analysis for activated Rho and total abundance of RhoA, -B, and -C, cyclin D1, and β-tubulin (loading control) in cyclin D1−/− MEFs transduced with vectors encoding wild-type cyclin D1 or vector control.
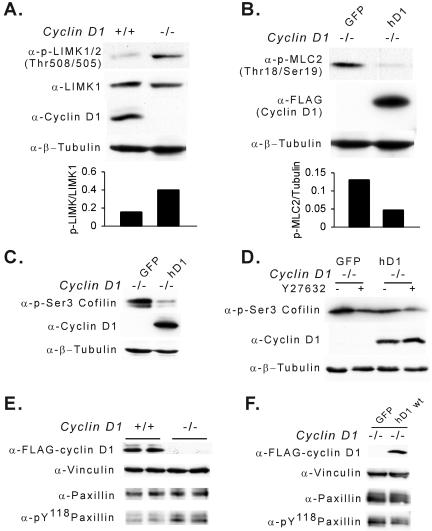
Substrates of the ROCK serine threonine kinase include LIM kinases at threonine 505/508 (48, 66), which regulates and phosphorylates the actin-depolymerizing protein cofilin at serine 3, and MLC2 (Thr18/Ser19) (33). To determine whether the increase in ROCKII abundance and kinase activity in cyclin D1−/− cells recapitulated activation of known ROCKII substrates, Western blot analysis was conducted with phosphor-specific antibodies to the ROCKII phosphorylation site of LIM kinase, MLC2, and cofilin. The phosphorylation of LIMK (Thr 505/508) was increased in cyclin D1−/− cells (Fig. 8A). The increase in LIMK phosphorylation was rescued by transduction with the cyclin D1 expression vector (data not shown). MLC2 and cofilin phosphorylation were decreased upon reexpression of cyclin D1 in cyclin D1−/− MEFs (Fig. 8B and C). Consistent with the biological importance of hyperactive ROCKII in the migratory defect of cyclin D1−/− MEFs (Fig. 7B and C) and the increased phosphorylation of cofilin by ROCKII in cyclin D1−/− MEFs, addition of the ROCK inhibitor Y27632 reduced the hyperphosphorylation of cofilin (Ser3) to the level of phosphorylation observed with cyclin D1 rescue (Fig. 8D, lane 2 versus lane 3). Consistent with the enhanced formation of focal contacts, the levels of tyrosine-phosphorylated paxillin (Y118) were increased in cyclin D1−/− cells (Fig. 8E) and reintroduction of cyclin D1 inhibited paxillin Y118 phosphorylation (Fig. 8F).
FIG. 8.
Cyclin D1 inhibits ROCK signaling. (A to D) Whole-cell lysates were examined by Western blotting for activation of ROCK signaling assessment using total and phosphorylated LIMK1, MLC2, and cofilin using MEFs from cyclin D1+/+, cyclin D1−/−, and cyclin D1−/− MEFs infected with MSCV-GFP virus control or MSCV-FLAG-tagged cyclin D1-IRES-GFP virus. α-, anti-. (E and F) Phosphorylated tyrosine 118 (pY118) paxillin assessed by Western blotting in cyclin D1+/+, cyclin D1−/−, and cyclin D1−/− MEFs infected with MSCV-GFP virus control or MSCV-FLAG-tagged cyclin D1-IRES-GFP virus.
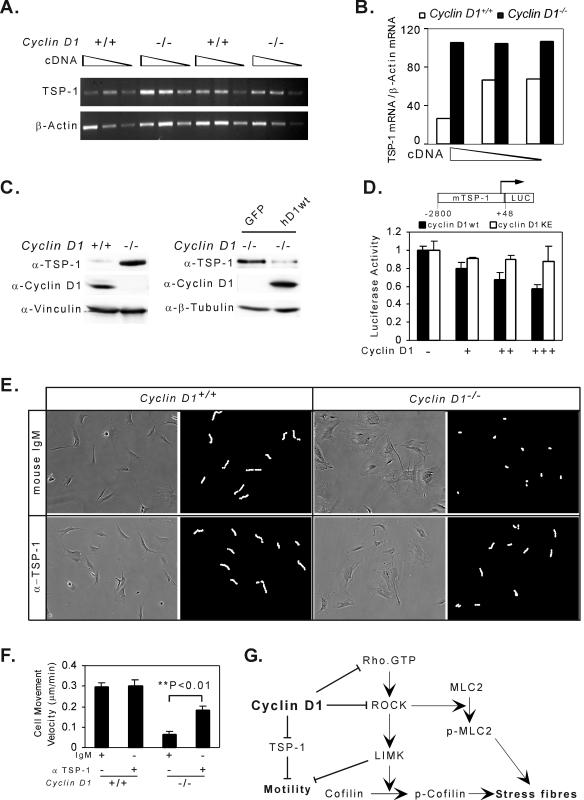
In view of the microarray analysis indicating an increase in TSP-1 abundance in each of the cyclin D1−/− MEFs analyzed (Table 1) and the known role for TSP-1 as an inhibitor of cellular migration, we examined further the relative abundance of TSP-1 and its regulation by cyclin D1. Expression of the angiogenesis inhibitor TSP-1, which is repressed by induction of the Ras-ROCK pathway (76), was induced in cyclin D1−/− MEFs using quantitative RT-PCR and Western blotting (Fig. 9A, B, and C). Reintroduction of cyclin D1 into cyclin D1−/− MEFs inhibited TSP-1 abundance (Fig. 9C). The murine TSP-1 promoter activity was repressed by cyclin D1 40 to 50% in a dose-dependent manner (Fig. 9D). The cdk-binding-defective mutant of cyclin D1, which failed to rescue the cyclin D1−/− migratory defect, failed to repress the TSP-1 promoter (Fig. 9D). In cyclin D1−/− MEFs, neutralizing TSP-1 antibody increased migration of cyclin D1−/− cells ∼3-fold compared with normal mouse IgM control (Fig. 9E and F). Addition of TSP-1 antibody to normal wild-type MEFs did not affect cellular migration, consistent with previous studies (72). The concurrent addition of the TSP-1 antibody to the Y27632-treated cyclin D1−/− cells provided no significant additional increase in cellular velocity compared with Y27632 alone (data not shown).
FIG. 9.
Cyclin D1 inhibits TSP-1 to promote cellular migration. (A and B) Semiquantitative RT-PCR of TSP-1 and β-actin from cyclin D1+/+ and cyclin D1−/− MEFs. (C) Western blot for TSP-1, cyclin D1, and vinculin (loading control) in cyclin D1+/+, cyclin D1−/−, and cyclin D1−/− MEFs infected with MSCV-GFP virus control or MSCV-FLAG-tagged cyclin D1-IRES-GFP virus. α-, anti-. (D) Murine TSP-1 promoter activity was repressed by wild-type cyclin D1. Luciferase reporter activity of murine TSP-1 promoter in the presence of increasing doses of cyclin D1 (wild type or KE mutant) expression vector in HEK 293 cells. Data are the mean + standard error for three separate transfections. (E) Cell movement velocity was determined by tracing single cells at 5-min intervals. Cell movement velocity was determined in the presence of either an immunoneutralizing antibody to TSP-1 or control IgM. (F) The immunoneutralizing TSP-1 antibody increased the cellular motility of cyclin D1−/− MEFs threefold (P < 0.01). (G) Model of cyclin D1 increasing cellular migration through the inhibition of TSP-1 and ROCK signaling.
Collectively, these studies show that cyclin D1 inhibits ROCKII expression and function as evidenced by cyclin D1-mediated inhibition of ROCKII expression and kinase activity, and the reduction in phosphorylation of LIMK1 (Thr508/505), MLC2 (Thr18/Ser19), and cofilin (Ser3) (Fig. 9G). In addition, cyclin D1 inhibits TSP-1 transcription and abundance. The increased production of TSP-1 by cyclin D1−/− cells contributes to the defect in cellular migration as reintroduction of cyclin D1 or immunoneutralizing antibody to TSP-1 reduced the migratory defect.
DISCUSSION
Herein, cyclin D1 promotes cellular migration and reduces cellular adhesion, independently of its pRb-binding function, but dependent upon an intact cdk-binding domain. Cyclin D1 reintroduction into cyclin D1−/− MEFs restored cellular morphology and migratory velocity. Microarray analysis identified ROCKII and TSP-1 as targets of cyclin D1-dependent repression. Immunoneutralizing antibodies to TSP-1 or addition of ROCKII inhibitor reversed the defective migration of cyclin D1-deficient cells. Together, these studies identify a novel mechanism by which cyclin D1 promotes cellular migration through inhibiting expression and function of ROCKII and TSP-1. The extracellular matrix glycoprotein TSP-1 inhibits cellular metastases and tumor growth (64). TSP-1 is a potent inhibitor of both tumorigenesis and in vivo neovascularization. Several oncogenes repress TSP-1, including oncogenic Ras, c-Myc, v-Src, and c-Jun (41, 51, 61, 67), as well as Id1 (72). Conversely, tumor suppressor proteins, including p53 and PTEN, induce TSP-1 abundance (17, 78). Ras repression of TSP-1 involves a signaling pathway, which includes the small monomeric GTPases and the Rho effectors (76). The current findings in which cyclin D1 reduced TSP-1 protein and mRNA abundance, repressing the TSP-1 promoter, are consistent with our previous findings that cyclin D1 is induced as a distal target of Ras, v-Src, and c-Jun (2, 35) and that each of these oncogenes represses TSP-1.
The current studies are consistent with recent findings linking factors that regulate cellular spreading and cell cycle progression. AP-1 proteins are known to promote both DNA synthesis and cellular motility (68, 71). Under several circumstances, cellular adhesion and migration promote G1-phase progression. Increased Rho activity, as observed in the cyclin D1−/− cells in the current studies, has been shown to natively regulate motility by increasing stress fiber-dependent adhesion (16). Although cellular adhesion is frequently required for cell cycle progression to occur, high levels of cell adhesion with large focal contacts results in failed cellular migration. Rho may also promote migration, being required for actomyosin contraction (4). Mutational analysis of the Rho/Rac chimera demonstrates distinct residues within the Rac effector domain regulate cellular morphology versus cyclin D1 expression and DNA synthesis (31, 80). Further evidence for a dissociation between the regulation of cellular morphology through Rho activity and DNA synthesis through pRb includes findings that unlike Rac and Cdc42, activated RhoA, despite its ability to induce morphological changes, does not inactivate pRb in NIH 3T3 cells (25). Together, these studies suggest the regulation of cell cycle progression and adhesion may be coupled or uncoupled, depending upon the cell type and signaling pathway involved.
Cyclin D1 promoted cell migration by inhibiting ROCKII expression and activity. ROCK kinase inhibitor reversed the defect in cellular motility in cyclin D1−/− cells, generating the more polarized morphology of wild-type cells. These findings are consistent with studies that ROCKII small interfering RNA induces an elongated morphology and that ROCK inhibitor treatment of cells induces a more polarized morphology (37) and increased cellular migration (6). Cyclin D1-deficient cells demonstrated evidence of increased ROCK activity using the in vitro substrate of myelin basic protein (MBP) and with increased phosphorylation of MLC2 (Thr18/Ser19), LIMK1/2 (Thr508/505), and cofilin (Ser3). The relationship between the activity of ROCK and cellular migration is complex and cell type specific. Thus, the ROCK inhibitor Y-27632 may either promote or inhibit cell migration (29, 46, 69, 81). The present findings are consistent with several recent findings. B-raf−/− MEFs demonstrated reduced ROCKII, reduced phosphocofilin, and increased migration (50), further confirming the correlation of increased ROCKII and reduced migration as seen in our studies. The possibility that the spread phenotype in cyclin D1−/− MEFs was a function of early senescence was excluded by the finding that pH-sensitive β-galactosidase staining (18) was similar between the cyclin D1−/− and cyclin D1+/+ MEFs used in the studies (data not shown).
Several lines of evidence suggest the reduced migration of cyclin D1 deficiency is not a direct consequence of reduced DNA synthesis. First, all cyclin D1−/− cells exhibited the migratory defect, whereas only 7% of the cells were in the DNA synthetic phase. Second, all cyclin D1−/− cells transduced by cyclin D1 demonstrated the change in cellular morphology, alterations in focal contact distribution and cellular migration, yet only a fraction of the cells were undergoing DNA synthesis. Third, the reintroduction of cyclin E through a knockin of the human cyclin E cDNA rescued the DNA synthetic defect of cyclin D1−/− cells but did not rescue either the migration defect nor the ROCKII abnormality. Collectively these studies suggest the effect of cyclin D1 on cell cycle progression and cellular migration may be dissociable functions. The effect of Rho, and its signaling components, including mDia, on cellular morphology can be uncoupled from effects on DNA synthesis. ROCK inhibition stimulates passage through G1 phase in the absence of cell spreading (37), and ROCK activity is not required for cell-shape-dependent G1 progression in microvascular endothelial cells. mDia, which promotes actin polymerization and regulates the alignment of stress fibers by targeting microtubules to focal adhesions, fails to promote G1-phase progression (37). Thus, interaction between ROCK and its regulation of cellular adhesion and DNA synthesis is dissociable and cell type dependent.
Cyclin D1 deficiency increased adhesion in both macrophages and MEFs, changes correlating with the induction of circumferential cortical F-actin stress fibers in MEFs and in macrophages (45). As with cyclin D1, genetic deletion of several other proteins known to regulate migration including RhoB, cyclin B, and p27kip1, do not appear to affect embryonic development (6, 10, 38, 40, 44, 49). The stable adhesive structures in cyclin D1-deficient cells are thought to contribute to the increased adhesion and reduced migration of these cells. It has been proposed that an increase in either Rac (40) or Rho (6) activity and/or their effectors can contribute to reduced cellular migration. Cyclin D1 transcription is induced by the Rho GTPases (12, 31, 79). Cyclin D1 induction in response to growth factors as a delayed early response at 6 h requires Rho-dependent sustained ERK activation and results in the induction of DNA synthesis. Rho also inhibits an alternative Rac/Cdc42-dependent induction of cyclin D1, thus preventing its premature induction (77). The inhibition of Rac-dependent expression of cyclin D1 involves LIM kinase through an effect that is independent of cofilin phosphorylation and actin polymerization. Herein, cyclin D1 also functions as an upstream inhibitor of Rho/ROCK/LIMK. As the effect of nuclear LIM kinase on cyclin D1 abundance regulates the duration of G1 phase (56), together these studies suggest cyclin D1 may function as a fine-tuning feedback-regulating LIM kinase.
Cyclin D1 physically interacts with pRb, p160 (AIB1), cdks, and the cell cycle inhibitor proteins p21CIP1 and p27KIP1. p21CIP1 promotes cell motility in Ras-transformed cells, through forming a complex with ROCK and thereby blocking Rho kinase action (36). p27KIP1 regulates actin dynamics, promoting cell migration (40, 43) independently of cyclin-cdk binding, suggesting that the mechanisms by which p27KIP1 and cyclin D1 regulate motility are distinct (40). p27KIP1 can function upstream of RhoA, inhibiting its activation (6). p21CIP1 and p27KIP1 can either reduce cdk activity or promote assembly and nuclear transport of D-type cyclins (59). Thus, new possible interactions between cyclin D1 and p27KIP1 in regulating cellular migration may require further analysis. The association of cyclin D1 overexpression with poor prognosis and tumor metastasis (19, 30) raised the intriguing possibility that cyclin D1 may play a distinct role in promoting cellular migration and invasion. The present studies demonstrate cyclin D1 promotes cellular migration through TSP-1 and ROCKII. The identification of compounds that selectively block the ATP pocket of cdk to selectively inhibit cellular kinase activity has proven challenging. The current findings that the cyclin D1 protein, through K112, promotes cellular migration may provide an important new avenue for therapy of metastatic disease in which cyclin D1 is overexpressed.
Acknowledgments
We are particularly grateful to P. Sicinski for the cyclin D1−/− and cyclin E knockin to cyclin D1−/− mice. We thank D. Scardino for assistance with manuscript preparation. We are grateful for the assistance of Susette C. Mueller (Microscopy and Imaging Shared Resource, Lombardi Cancer Center, Georgetown University) for technical advice.
Support for these studies was from NIH R01CA70896, R01CA75503, R01CA86072, R01CA93596, R01CA107382, and Kimmel Cancer Center Support grant P30 CA56036 to R.G.P.; CA26504, P01 100324-02, and P30 CA13330-31 to E.R.S.; and the Breast Cancer Alliance through an Exceptional Project grant to A.A.Q.
REFERENCES
- 1.Albanese, C., M. D'Amico, A. T. Reutens, M. Fu, G. Watanabe, R. J. Lee, R. N. Kitsis, B. Henglein, M. Avantaggiati, K. Somasundaram, B. Thimmapaya, and R. G. Pestell. 1999. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J. Biol. Chem. 274:34186-34195. [DOI] [PubMed] [Google Scholar]
- 2.Albanese, C., J. Johnson, G. Watanabe, N. Eklund, D. Vu, A. Arnold, and R. G. Pestell. 1995. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 270:23589-23597. [DOI] [PubMed] [Google Scholar]
- 3.Albanese, C., K. Wu, M. D'Amico, C. Jarrett, D. Joyce, J. Hughes, J. Hulit, T. Sakamaki, M. Fu, A. Ben-Ze'ev, J. F. Bromberg, C. Lamberti, U. Verma, R. B. Gaynor, S. W. Byers, and R. G. Pestell. 2003. IKKalpha regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Mol. Biol. Cell 14:585-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen, W. E., G. E. Jones, J. W. Pollard, and A. J. Ridley. 1997. Rho, Rac and Cdc42 regulate actin organization and cell adhesion in macrophages. J. Cell Sci. 110:707-720. [DOI] [PubMed] [Google Scholar]
- 5.Amano, M., K. Chihara, K. Kimura, Y. Fukata, N. Nakamura, Y. Matsuura, and K. Kaibuchi. 1997. Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science 275:1308-1311. [DOI] [PubMed] [Google Scholar]
- 6.Besson, A., M. Gurian-West, A. Schmidt, A. Hall, and J. M. Roberts. 2004. p27Kip1 modulates cell migration through the regulation of RhoA activation. Genes Dev. 18:862-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bornstein, P., D. Alfi, S. Devarayalu, P. Framson, and P. Li. 1990. Characterization of the mouse thrombospondin gene and evaluation of the role of the first intron in human gene expression. J. Biol. Chem. 265:16691-16698. [PubMed] [Google Scholar]
- 8.Bouras, T., M. Fu, A. A. Sauve, F. Wang, A. A. Quong, N. D. Perkins, R. T. Hay, W. Gu, and R. G. Pestell. 2005. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J. Biol. Chem. 280:10264-10276. [DOI] [PubMed] [Google Scholar]
- 9.Boutros, P. C., I. D. Moffat, M. A. Franc, N. Tijet, J. Tuomisto, R. Pohjanvirta, and A. B. Okey. 2004. Dioxin-responsive AHRE-II gene battery: identification by phylogenetic footprinting. Biochem. Biophys. Res. Commun. 321:707-715. [DOI] [PubMed] [Google Scholar]
- 10.Brandeis, M., I. Rosewell, M. Carrington, T. Crompton, M. A. Jacobs, J. Kirk, J. Gannon, and T. Hunt. 1998. Cyclin B2-null mice develop normally and are fertile whereas cyclin B1-null mice die in utero. Proc. Natl. Acad. Sci. USA 95:4344-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bromberg, J. F., M. H. Wrzeszczynska, G. Devgan, Y. Zhao, R. G. Pestell, C. Albanese, and J. E. Darnell. 1999. Stat3 as an oncogene. Cell 98:295-303. [DOI] [PubMed] [Google Scholar]
- 12.Burbelo, P., A. Wellstein, and R. G. Pestell. 2004. Altered Rho GTPase signaling pathways in breast cancer cells. Breast Cancer Res. Treat. 84:43-48. [DOI] [PubMed] [Google Scholar]
- 13.Burridge, K., and K. Wennerberg. 2004. Rho and Rac take center stage. Cell 116:167-179. [DOI] [PubMed] [Google Scholar]
- 14.Chen, Y., V. Kamat, E. R. Dougherty, M. L. Bittner, P. S. Meltzer, and J. M. Trent. 2002. Ratio statistics of gene expression levels and applications to microarray data analysis. Bioinformatics 18:1207-1215. [DOI] [PubMed] [Google Scholar]
- 15.Cox, D., J. S. Berg, M. Cammer, J. O. Chinegwundoh, B. M. Dale, R. E. Cheney, and S. Greenberg. 2002. Myosin X is a downstream effector of PI(3)K during phagocytosis. Nat. Cell Biol. 4:469-477. [DOI] [PubMed] [Google Scholar]
- 16.Cox, E. A., S. K. Sastry, and A. Huttenlocher. 2001. Integrin-mediated adhesion regulates cell polarity and membrane protrusion through the Rho family of GTPases. Mol. Biol. Cell 12:265-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dameron, K. M., O. V. Volpert, M. A. Tainsky, and N. Bouck. 1994. Control of angiogenesis in fibroblasts by p53 regulation of thrombospondin-1. Science 265:1582-1584. [DOI] [PubMed] [Google Scholar]
- 18.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drobnjak, M., I. Osman, H. I. Scher, M. Fazzari, and C. Cordon-Cardo. 2000. Overexpression of cyclin D1 is associated with metastatic prostate cancer to bone. Clin. Cancer Res. 6:1891-1895. [PubMed] [Google Scholar]
- 20.Fahraeus, R., S. Lain, K. L. Ball, and D. P. Lane. 1998. Characterization of the cyclin-dependent kinase inhibitory domain of the INK4 family as a model for a synthetic tumour suppressor molecule. Oncogene 16:587-596. [DOI] [PubMed] [Google Scholar]
- 21.Fahraeus, R., and D. P. Lane. 1999. The p16(INK4a) tumour suppressor protein inhibits alphavbeta3 integrin-mediated cell spreading on vitronectin by blocking PKC-dependent localization of alphavbeta3 to focal contacts. EMBO J. 18:2106-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu, M., M. Rao, T. Bouras, C. Wang, K. Wu, X. Zhang, Z. Li, T. P. Yao, and R. G. Pestell. 2005. Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. J. Biol. Chem. 280:16934-16941. [DOI] [PubMed] [Google Scholar]
- 23.Fu, M., C. Wang, Z. Li, T. Sakamaki, and R. G. Pestell. 2004. Minireview. Cyclin D1: normal and abnormal functions. Endocrinology 145:5439-5447. [DOI] [PubMed] [Google Scholar]
- 24.Geng, Y., W. Whoriskey, M. Y. Park, R. T. Bronson, R. H. Medema, T. Li, R. A. Weinberg, and P. Sicinski. 1999. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell 97:767-777. [DOI] [PubMed] [Google Scholar]
- 25.Gjoerup, O., J. Lukas, J. Bartek, and B. M. Willumsen. 1998. Rac and Cdc42 are potent stimulators of E2F-dependent transcription capable of promoting retinoblastoma susceptibility gene product hyperphosphorylation. J. Biol. Chem. 273:18812-18818. [DOI] [PubMed] [Google Scholar]
- 26.Good, D. J., P. J. Polverini, F. Rastinejad, M. M. Le Beau, R. S. Lemons, W. A. Frazier, and N. P. Bouck. 1990. A tumor suppressor-dependent inhibitor of angiogenesis is immunologically and functionally indistinguishable from a fragment of thrombospondin. Proc. Natl. Acad. Sci. USA 87:6624-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosotani, R., Y. Miyamoto, K. Fujimoto, R. Doi, A. Otaka, N. Fujii, and M. Imamura. 2002. Trojan p16 peptide suppresses pancreatic cancer growth and prolongs survival in mice. Clin. Cancer Res. 8:1271-1276. [PubMed] [Google Scholar]
- 28.Hulit, J., C. Wang, Z. Li, C. Albanese, M. Rao, D. Di Vizio, S. Shah, S. W. Byers, R. Mahmood, L. H. Augenlicht, R. Russell, and R. G. Pestell. 2004. Cyclin D1 genetic heterozygosity regulates colonic epithelial cell differentiation and tumor number in ApcMin mice. Mol. Cell. Biol. 24:7598-7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itoh, K., K. Yoshioka, et al. 1999. An essential part for Rho-associated kinase in the transcellular invasion of tumor cells. Nat. Med. 5:221-225. [DOI] [PubMed] [Google Scholar]
- 30.Jares, P., P. L. Fernandez, E. Campo, A. Nadal, F. Bosch, G. Aiza, I. Nayach, J. Traserra, and A. Cardesa. 1994. PRAD-1/cyclin D1 gene amplification correlates with messenger RNA overexpression and tumor progression in human laryngeal carcinomas. Cancer Res. 54:4813-4817. [PubMed] [Google Scholar]
- 31.Joyce, D., B. Bouzahzah, M. Fu, C. Albanese, M. D'Amico, J. Steer, J. U. Klein, R. J. Lee, J. E. Segall, J. K. Westwick, C. J. Der, and R. G. Pestell. 1999. Integration of Rac-dependent regulation of cyclin D1 transcription through an NF-κB-dependent pathway. J. Biol. Chem. 274:25245-25249. [DOI] [PubMed] [Google Scholar]
- 32.Kimura, K., M. Ito, M. Amano, K. Chihara, Y. Fukata, M. Nakafuku, B. Yamamori, J. Feng, T. Nakano, K. Okawa, A. Iwamatsu, and K. Kaibuchi. 1996. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273:245-248. [DOI] [PubMed] [Google Scholar]
- 33.Kureishi, Y., S. Kobayashi, M. Amano, K. Kimura, H. Kanaide, T. Nakano, K. Kaibuchi, and M. Ito. 1997. Rho-associated kinase directly induces smooth muscle contraction through myosin light chain phosphorylation. J. Biol. Chem. 272:12257-12260. [DOI] [PubMed] [Google Scholar]
- 34.Lee, R. J., C. Albanese, M. Fu, M. D'Amico, B. Lin, G. Watanabe, G. K. Haines III, P. M. Siegel, M.-C. Hung, Y. Yarden, J. M. Horowitz, W. J. Muller, and R. G. Pestell. 2000. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol. Cell. Biol. 20:672-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, R. J., C. Albanese, R. J. Stenger, G. Watanabe, G. Inghirami, G. K. I. Haines, M. Webster, W. J. Muller, J. S. Brugge, R. J. Davis, and R. G. Pestell. 1999. pp60v-src induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways: a role for cAMP response element-binding protein and activating transcription factor-2 in pp60v-src signaling in breast cancer cells. J. Biol. Chem. 274:7341-7350. [DOI] [PubMed] [Google Scholar]
- 36.Lee, S., and D. M. Helfman. 2004. Cytoplasmic p21Cip1 is involved in Ras-induced inhibition of the ROCK/LIMK/cofilin pathway. J. Biol. Chem. 279:1885-1891. [DOI] [PubMed] [Google Scholar]
- 37.Mammoto, A., S. Huang, K. Moore, P. Oh, and D. E. Ingber. 2004. Role of RhoA, mDia, and ROCK in cell shape-dependent control of the Skp2-p27kip1 pathway and the G1/S transition. J. Biol. Chem. 279:26323-26330. [DOI] [PubMed] [Google Scholar]
- 38.Manes, T., D. Q. Zheng, S. Tognin, A. S. Woodard, P. C. Marchisio, and L. R. Languino. 2003. Alpha(v)beta3 integrin expression up-regulates cdc2, which modulates cell migration. J. Cell Biol. 161:817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massague, J. 2004. G1 cell-cycle control and cancer. Nature 432:298-306. [DOI] [PubMed] [Google Scholar]
- 40.McAllister, S. S., M. Becker-Hapak, G. Pintucci, M. Pagano, and S. F. Dowdy. 2003. Novel p27kip1 C-terminal scatter domain mediates Rac-dependent cell migration independent of cell cycle arrest functions. Mol. Cell. Biol. 23:216-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mettouchi, A., F. Cabon, N. Montreau, P. Vernier, G. Mercier, D. Blangy, H. Tricoire, P. Vigier, and B. Binetruy. 1994. SPARC and thrombospondin genes are repressed by the c-jun oncogene in rat embryo fibroblasts. EMBO J. 13:5668-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motokura, T., T. Bloom, H. G. Kim, H. Jüppner, J. V. Ruderman, H. M. Kronenberg, and A. Arnold. 1991. A novel cyclin encoded by a bcl1-linked candidate oncogene. Nature 350:512-515. [DOI] [PubMed] [Google Scholar]
- 43.Nagahara, H., A. M. Vocero-Akbani, E. L. Snyder, A. Ho, D. G. Latham, N. A. Lissy, M. Becker-Hapak, S. A. Ezhevsky, and S. F. Dowdy. 1998. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat. Med. 4:1449-1452. [DOI] [PubMed] [Google Scholar]
- 44.Nakayama, K., N. Ishida, M. Shirane, A. Inomata, T. Inoue, N. Shishido, I. Horii, D. Y. Loh, and K.-I. Nakayama. 1996. Mice lacking p27Kip1 display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85:707-720. [DOI] [PubMed] [Google Scholar]
- 45.Neumeister, P., F. J. Pixley, Y. Xiong, H. Xie, K. Wu, A. Ashton, M. Cammer, A. Chan, M. Symons, E. R. Stanley, and R. G. Pestell. 2003. Cyclin D1 governs adhesion and motility of macrophages. Mol. Biol. Cell 14:2005-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nobes, C. D., and A. Hall. 1999. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 144:1235-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oh, E.-S., H. Gu, T. M. Saxton, J. F. Timms, S. Hausdorff, E. U. Frevert, B. B. Kahn, T. Pawson, B. G. Neel, and S. M. Thomas. 1999. Regulation of early events in integrin signaling by protein tyrosine phosphatase SHP-2. Mol. Cell. Biol. 19:3205-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohashi, K., K. Nagata, M. Maekawa, T. Ishizaki, S. Narumiya, and K. Mizuno. 2000. Rho-associated kinase ROCK activates LIM-kinase 1 by phosphorylation at threonine 508 within the activation loop. J. Biol. Chem. 275:3577-3582. [DOI] [PubMed] [Google Scholar]
- 49.Prendergast, G. C. 2001. Actin' up: RhoB in cancer and apoptosis. Nat. Rev. Cancer 1:162-168. [DOI] [PubMed] [Google Scholar]
- 50.Pritchard, C. A., L. Hayes, L. Wojnowski, A. Zimmer, R. M. Marais, and J. C. Norman. 2004. B-Raf acts via the ROCKII/LIMK/cofilin pathway to maintain actin stress fibers in fibroblasts. Mol. Cell. Biol. 24:5937-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rak, J., Y. Mitsuhashi, C. Sheehan, A. Tamir, A. Viloria-Petit, J. Filmus, S. J. Mansour, N. G. Ahn, and R. S. Kerbel. 2000. Oncogenes and tumor angiogenesis: differential modes of vascular endothelial growth factor up-regulation in ras-transformed epithelial cells and fibroblasts. Cancer Res. 60:490-498. [PubMed] [Google Scholar]
- 52.Ren, X. D., W. B. Kiosses, D. J. Sieg, C. A. Otey, D. D. Schlaepfer, and M. A. Schwartz. 2000. Focal adhesion kinase suppresses Rho activity to promote focal adhesion turnover. J. Cell Sci. 113:3673-3678. [DOI] [PubMed] [Google Scholar]
- 53.Renn, S. C., N. Aubin-Horth, and H. A. Hofmann. 2004. Biologically meaningful expression profiling across species using heterologous hybridization to a cDNA microarray. BMC Genomics 5:42. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ridley, A. J., and A. Hall. 1992. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70:389-399. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez-Manzaneque, J. C., T. F. Lane, M. A. Ortega, R. O. Hynes, J. Lawler, and M. L. Iruela-Arispe. 2001. Thrombospondin-1 suppresses spontaneous tumor growth and inhibits activation of matrix metalloproteinase-9 and mobilization of vascular endothelial growth factor. Proc. Natl. Acad. Sci. USA 98:12485-12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roovers, K., E. A. Klein, P. Castagnino, and R. K. Assoian. 2003. Nuclear translocation of LIM kinase mediates Rho-Rho kinase regulation of cyclin D1 expression. Dev. Cell 5:273-284. [DOI] [PubMed] [Google Scholar]
- 57.Sahai, E., and C. J. Marshall. 2003. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 5:711-719. [DOI] [PubMed] [Google Scholar]
- 58.Sahai, E., M. F. Olson, and C. J. Marshall. 2001. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 20:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 60.Sicinski, P., J. L. Donaher, S. B. Parker, T. Li, A. Fazeli, H. Gardner, S. Z. Haslam, R. T. Bronson, S. J. Elledge, and R. A. Weinberg. 1995. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82:621-630. [DOI] [PubMed] [Google Scholar]
- 61.Slack, J. L., and P. Bornstein. 1994. Transformation by v-src causes transient induction followed by repression of mouse thrombospondin-1. Cell Growth Differ. 5:1373-1380. [PubMed] [Google Scholar]
- 62.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article 3. [Online.] http://www.bepress.com/sagmb/vol3/iss1/art3. [DOI] [PubMed]
- 63.Stellmach, V., O. V. Volpert, S. E. Crawford, J. Lawler, R. O. Hynes, and N. Bouck. 1996. Tumour suppressor genes and angiogenesis: the role of TP53 in fibroblasts. Eur. J. Cancer 32A:2394-2400. [DOI] [PubMed] [Google Scholar]
- 64.Streit, M., P. Velasco, L. F. Brown, M. Skobe, L. Richard, L. Riccardi, J. Lawler, and M. Detmar. 1999. Overexpression of thrombospondin-1 decreases angiogenesis and inhibits the growth of human cutaneous squamous cell carcinomas. Am. J. Pathol. 155:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Streit, M., P. Velasco, L. Riccardi, L. Spencer, L. F. Brown, L. Janes, B. Lange-Asschenfeldt, K. Yano, T. Hawighorst, L. Iruela-Arispe, and M. Detmar. 2000. Thrombospondin-1 suppresses wound healing and granulation tissue formation in the skin of transgenic mice. EMBO J. 19:3272-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sumi, T., K. Matsumoto, and T. Nakamura. 2001. Specific activation of LIM kinase 2 via phosphorylation of threonine 505 by ROCK, a Rho-dependent protein kinase. J. Biol. Chem. 276:670-676. [DOI] [PubMed] [Google Scholar]
- 67.Tikhonenko, A. T., D. J. Black, and M. L. Linial. 1996. Viral Myc oncoproteins in infected fibroblasts down-modulate thrombospondin-1, a possible tumor suppressor gene. J. Biol. Chem. 271:30741-30747. [DOI] [PubMed] [Google Scholar]
- 68.Tkach, V., E. Tulchinsky, E. Lukanidin, C. Vinson, E. Bock, and V. Berezin. 2003. Role of the Fos family members, c-Fos, Fra-1 and Fra-2, in the regulation of cell motility. Oncogene 22:5045-5054. [DOI] [PubMed] [Google Scholar]
- 69.Totsukawa, G., Y. Wu, Y. Sasaki, D. J. Hartshorne, Y. Yamakita, S. Yamashiro, and F. Matsumura. 2004. Distinct roles of MLCK and ROCK in the regulation of membrane protrusions and focal adhesion dynamics during cell migration of fibroblasts. J. Cell Biol. 164:427-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uehata, M., T. Ishizaki, H. Satoh, T. Ono, T. Kawahara, T. Morishita, H. Tamakawa, K. Yamagami, J. Inui, M. Maekawa, and S. Narumiya. 1997. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature 389:990-994. [DOI] [PubMed] [Google Scholar]
- 71.Vial, E., E. Sahai, and C. J. Marshall. 2003. ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell 4:67-79. [DOI] [PubMed] [Google Scholar]
- 72.Volpert, O. V., R. Pili, H. A. Sikder, T. Nelius, T. Zaichuk, C. Morris, C. B. Shiflett, M. K. Devlin, K. Conant, and R. M. Alani. 2002. Id1 regulates angiogenesis through transcriptional repression of thrombospondin-1. Cancer Cell 2:473-483. [DOI] [PubMed] [Google Scholar]
- 73.Wang, C., Z. Li, M. Fu, T. Bouras, and R. G. Pestell. 2004. Signal transduction mediated by cyclin D1: from mitogens to cell proliferation: a molecular target with therapeutic potential. Cancer Treat. Res. 119:217-237. [DOI] [PubMed] [Google Scholar]
- 74.Wang, C., N. Pattabiraman, J. N. Zhou, M. Fu, T. Sakamaki, C. Albanese, Z. Li, K. Wu, J. Hulit, P. Neumeister, P. M. Novikoff, M. Brownlee, P. E. Scherer, J. G. Jones, K. D. Whitney, L. A. Donehower, E. L. Harris, T. Rohan, D. C. Johns, and R. G. Pestell. 2003. Cyclin D1 repression of peroxisome proliferator-activated receptor γ expression and transactivation. Mol. Cell. Biol. 23:6159-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watanabe, G., R. J. Lee, C. Albanese, W. E. Rainey, D. Batlle, and R. G. Pestell. 1996. Angiotensin II (AII) activation of cyclin D1-dependent kinase activity. J. Biol. Chem. 271:22570-22577. [DOI] [PubMed] [Google Scholar]
- 76.Watnick, R. S., Y. N. Cheng, A. Rangarajan, T. A. Ince, and R. A. Weinberg. 2003. Ras modulates Myc activity to repress thrombospondin-1 expression and increase tumor angiogenesis. Cancer Cell 3:219-231. [DOI] [PubMed] [Google Scholar]
- 77.Welsh, C. F., K. Roovers, J. Villanueva, Y. Liu, M. A. Schwartz, and R. K. Assoian. 2001. Timing of cyclin D1 expression within G1 phase is controlled by Rho. Nat. Cell Biol. 3:950-957. [DOI] [PubMed] [Google Scholar]
- 78.Wen, S., J. Stolarov, M. P. Myers, J. D. Su, M. H. Wigler, N. K. Tonks, and D. L. Durden. 2001. PTEN controls tumor-induced angiogenesis. Proc. Natl. Acad. Sci. USA 98:4622-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Westwick, J. K., Q. T. Lambert, G. J. Clark, M. Symons, L. Van Aelst, R. G. Pestell, and C. J. Der. 1997. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol. Cell. Biol. 17:1324-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Westwick, J. K., R. J. Lee, Q. T. Lambert, M. Symons, R. G. Pestell, C. J. Der, and I. P. Whitehead. 1998. Transforming potential of Dbl family proteins correlates with transcription from the cyclin D1 promoter but not with activation of Jun NH2-terminal kinase, p38/Mpk2, serum response factor, or c-Jun. J. Biol. Chem. 273:16739-16747. [DOI] [PubMed] [Google Scholar]
- 81.Wojciak-Stothard, B., and A. J. Ridley. 2003. Shear stress-induced endothelial cell polarization is mediated by Rho and Rac but not Cdc42 or PI 3-kinases. J. Cell Biol. 161:429-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]