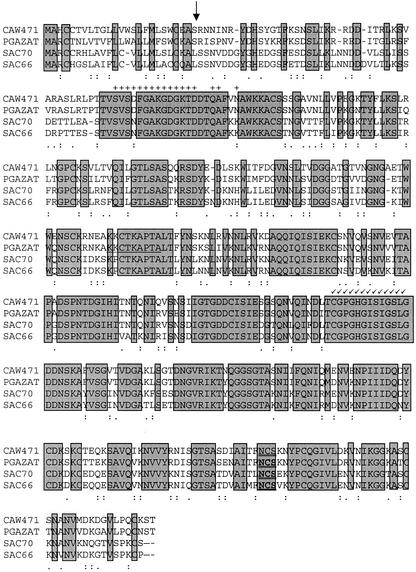
Figure 1.
Comparison of the predicted peptide sequence of CAW471 with that of PGAZAT, SAC70 (Jenkins et al., 1999), and SAC66 (Jenkins et al., 1999). Identical residues are in shaded boxes; a colon represents similar residues, whereas a period represents related residues. The predicted signal peptide cleavage site is indicated by an arrow (↓) and the conserved Asn glycosylation site (N-X-S/T) is double underlined. The exon previously omitted from the PGAZAT sequence is underlined and conserved residues in other endo- and exo-PGs are indicated by + and √, respectively. The alignment was generated using a Clustal method using the Pam 250 residue weight table on the DNAstar program. Dark shading indicates identical residues.