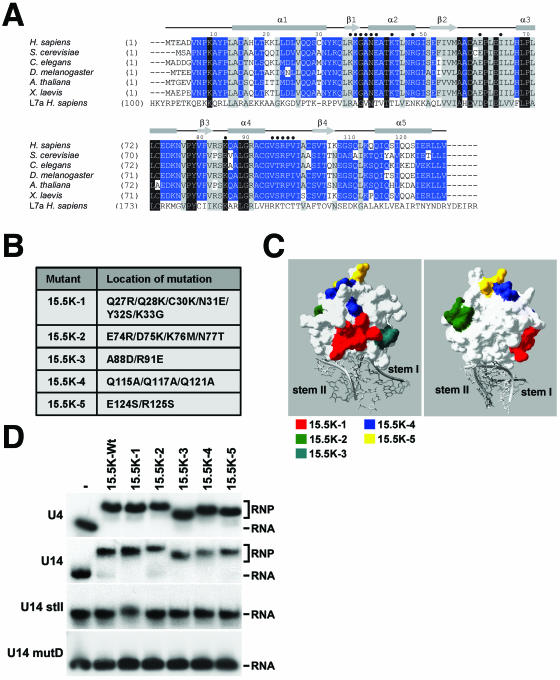
FIG. 2.
Identification and mutation of conserved amino acids on the surface of 15.5K. A) Amino acid alignment of 15.5K from human (Homo sapiens), Saccharomyces cerevisiae (Snu13p), Caenorhabditis elegans (accession no. Q21568), Drosophila melanogaster (accession no. GH03082), Arabidopsis thaliana (accession no. A71421), and Xenopus laevis (accession no. AAH46579) with the human ribosomal protein L7a sequence using the ClustalV program. Conserved residues are shown on a gray background and are grouped as described by Schulz and Schirmer (24). Identical and conserved residues are indicated by white type on black background and by black type on gray background, respectively. Residues specifically conserved in 15.5K are depicted in white type on blue background. Amino acid positions are indicated on the left. The secondary structure of human 15.5K (29) is indicated above the alignment, and amino acids involved in RNA binding are indicated by dark blue circles. B) The five mutations introduced into 15.5K. C) The positions of the residues targeted for mutagenesis on the surface of 15.5K are depicted using the crystal structure of 15.5K bound to the 5′ stem-loop of the U4 snRNA (29). The surface plot of 15.5K is shown in white, and the U4 snRNA is shaded gray. The positions of the amino acids targeted for mutagenesis are indicated in either red (mutant 15.5K-1), green (15.5K-2), turquoise (15.5K-3), blue (15.5K-4), or yellow (15.5K-5). D) Gel mobility shift analysis of the interaction of recombinant 15.5K and 15.5K mutants with U4, U14 and U14 mutants. The mutant proteins outlined in panel B were incubated with 32P-labeled in vitro-transcribed snRNA or snoRNA transcripts, and the resulting RNA-protein complexes were resolved on a 6% native polyacrylamide gel and visualized by autoradiography. The identity of the protein used is indicated above each lane (−, no 15.5K; 15.5K-Wt, wild-type 15.5K). The positions of the protein-RNA complex (RNP) and the free RNA are indicated on the right. The RNA used is indicated on the left.