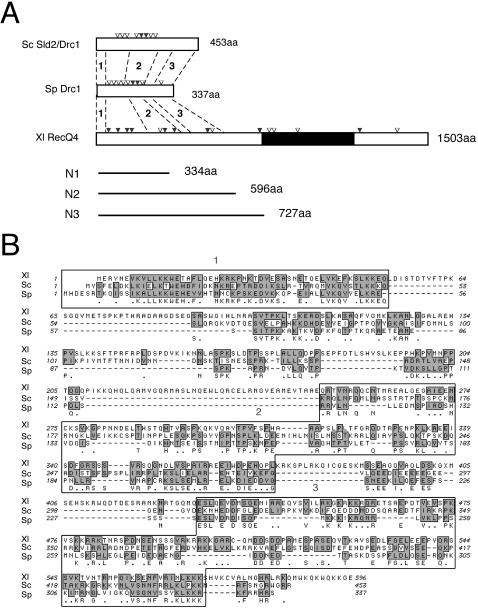
FIG. 1.
Sequence comparison of Sld2, Drc1, and RecQ4. (A) Schematic structures of Sld2/Drc1 of Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), and Xenopus laevis (Xl) RecQ4. Potential CDK phosphorylation motifs (S/T-P, S/T-P-K/R, and S/T-P-X-K/R) are indicated by open (one motif) and solid (two motifs) inverted triangles. The black region represents the RecQ helicase domain (amino acids 744 to 1086). Bold lines represent N-terminal fragments of Xenopus RecQ4 used in this study. Regions 1, 2, and 3 show the following identity and similarity (% identity/% similarity): Xl RecQ4/Sc Sld2, 30/55, 19/31, and 19/35; Xl RecQ4/Sp Drc1, 35/52, 16/28, and 23/34; and Sc Sld2/Sp Drc1, 29/55, 26/40, and 20/32, respectively. (B) Alignment of N-terminal amino acid sequences of Sc Sld2/Drc1, Sp Drc1, and Xl RecQ4. Identical amino acids are indicated just below the sequence of Sp Drc1, and conserved regions are shaded. Boxed regions show relatively high sequence similarities.