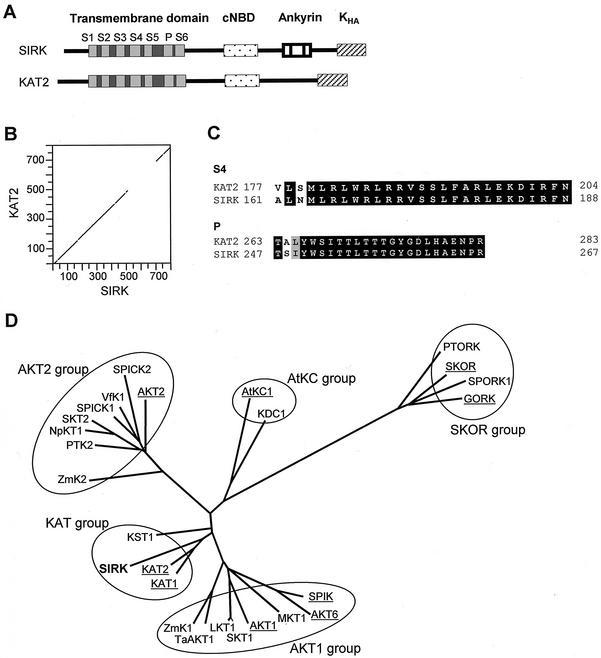
Figure 1.
The grapevine SIRK gene encodes a Shaker-like K+ channel related to the Arabidopsis KAT2 channel. A, Schematic representations of the predicted domains of SIRK and KAT2 channels. S1 through S6, Six membrane-spanning segments (light gray) and linkers (dark gray) forming the channel transmembrane domain. P, Conserved pore-forming domain. KHA, C-terminal domain supposed to be involved in channel subunit tetramerization and/or clustering in the membrane. B, Dot matrix comparison (DNA Strider program [Marck, 1990] polypeptide homology matrix; stringency = 7, window = 15 amino acids) of the deduced SIRK amino acid sequence (accession no. AF359521) with that of KAT2 (accession no. AJ288900). C, Alignment of SIRK and KAT2 voltage sensor (S4) and pore sequences. Identical amino acids are written in white, similar amino acids are written in black on a gray background. D, Phylogenetic tree of plant potassium channels deduced from full-length cDNA. This tree was generated by the Darwin program (Gonnet et al., 1992) with the complete protein sequences of the channels. Arabidopsis sequences are underlined. Protein accession numbers are AKT1, S23606; SKT1, T07651; ZmK1, T03939; SPIK, AJ309323; AKT6, CAA22577; LKT1, X96390; KAT1, S32816; KAT2, T04931; KST1, S55349; AKT2, AAA97865; NpKT1, BAA84085; SKT2, T07052; VfK1, T12177; SPICK1, AAD16278; SPICK2, AAD39492; ZmK2, CAB54856; AtKC1, G7488024; KDC1, AJ249962; SKOR, CAA11280; TaAKT1, AF207745; GORK, AJ279009; SPORK1, AJ299019; PTK2, AJ271447; PTORK, AJ271446; MKT1, AF267753; and SIRK, AF359521.