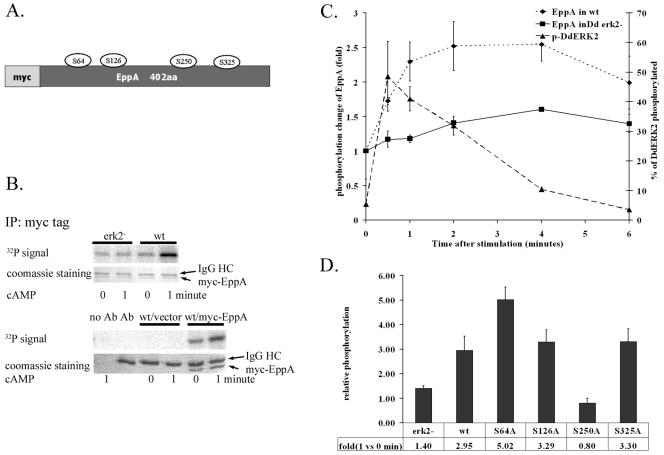
FIG. 3.
ERK2-mediated phosphorylation of EppA. (A) myc-tagged EppA construct and putative phosphorylation sites that were mutated. (B) ERK2-dependent in vivo phosphorylation of myc-EppA. (Upper panel) myc-EppA was immunoprecipitated from lysates of 32P-labeled wild-type (wt) or Dderk2− (erk2−) Dictyostelium cells stably expressing myc-EppA before and after stimulation with 10 μM cAMP. Immunoprecipitates (IP) were separated by SDS-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue, and phosphorylation was quantitated by a PhosphorImager. (Lower panel) Control to confirm that the lower band is myc-EppA. IgG, immunoglobulin G. (C) Time course of cAMP-induced EppA phosphorylation. Wild-type and Dderk2− cells overexpressing myc-tagged EppA were starved 6 h and labeled with 32PO43−. myc-tagged EppA was immunoprecipitated from lysates prepared at different time points after cAMP stimulation. The phosphorylation level at each time point was standardized by the amount of myc-EppA. Phosphorylation of DdERK2 after cAMP stimulation was detected by Western blotting, and the densities of corresponding bands were measured and used to determine the percentage of DdERK2 phosphorylated. Values are averages from two experiments. Error bars indicate the range of the original data. (D) Identification of phosphorylation sites. Cells stably expressing myc-EppA constructs carrying the single point mutations identified in panel A were labeled and stimulated, and the myc-tagged protein was immunoprecipitated for SDS-polyacrylamide gel electrophoretic analysis as for panel B. The 32P signal was normalized to myc-EppA protein staining, and the relative change in EppA phosphorylation (32P density of stimulated sample/density of unstimulated sample) was averaged from three to four experiments.