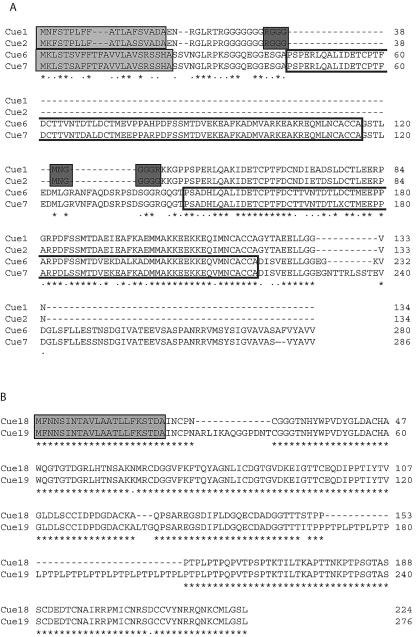
FIG. 2.
Amino acid sequence comparisons of predicted Cue proteins located within an inverted repeat on genomic scaffold 73. Alignments were performed with CLUSTAL W (1.81) (54). Sequences are identified on the left, and amino acid residue numbers are indicated on the right. Dashes in sequences were introduced to facilitate maximum alignment. Below the sequences, asterisks denote amino acid residues conserved in all sequences and dots indicate conservative replacements. Signal sequences were predicted with SignalP 3.0 (6) and are enclosed within light gray boxes. (A) Alignment of Cue1, Cue2, Cue6, Cue7. The repeated domain in Cue6 and Cue7 is encompassed within unshaded boxes. PredictNLS (10) identified a potential NLS in Cue1 and Cue2, denoted by dark gray boxes. (B) Alignment of Cue18 and Cue19.