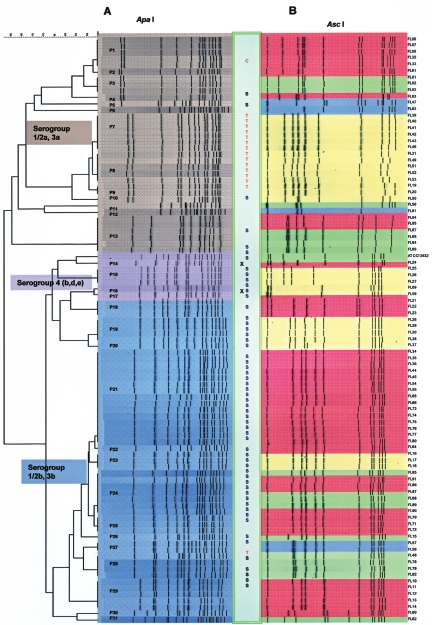
FIG. 1.
Cluster analysis of L. monocytogenes isolates along with phenotypic characteristics. PFGE was performed after genomic DNA was digested with ApaI (A) or AscI (B) using the CDC PulseNet standardized procedure (11). Digestion patterns for both restriction enzymes were used to determine PFGE types (PFGE types P1 to P31 in panel A). PCR-based serotype groups are indicated by brown [serotype group 1/2a(3a)], violet [serotype group 4b(d,e)], and blue [serotype group 1/2b(3b)]. The box between the two panels indicates antibiotic resistance (T, tetracycline; C, carbenicillin; S, sulfomethoxazole) and an inability to induce glutamate-dependent acid resistance under aerobic conditions (X). Susceptibility to the listeriophage cocktail was expressed as the number of PFU generated per ml by individual isolates and is indicated as follows: yellow, 0 to 101 PFU/ml; blue, 102 to 104 PFU/ml; green, 105 to 106 PFU/ml; and red, >107 PFU/ml.