Abstract
MgtC is a virulence factor common to several intracellular pathogens, including Mycobacterium tuberculosis, that might have been acquired through horizontal gene transfer. In the present study, we investigated the polymorphism of mgtC in clinical isolates representative of the main epidemic groups of M. tuberculosis. MgtC appears to have a low polymorphism rate in M. tuberculosis that consists exclusively of nonsynonymous mutations. We identified a single nucleotide polymorphism (SNP) at mgtC codon 182 (mgtC182) specifically associated with the Haarlem genotype. A simple PCR assay, called the “on/off switch assay,” using phosphorothioate-modified primers and Pfu polymerase allowed us to distinguish Haarlem from non-Haarlem strains based on the mgtC182 SNP. The amino acid change (H182R) associated with the mgtC182 SNP in Haarlem strains does not appear to procure a selective advantage. Our results offer a simple and rapid tool to distinguish between Haarlem and non-Haarlem strains. In addition, the on/off switch assay, which allows the detection of SNPs on chromosomal DNA and M. tuberculosis cultures, provides a novel approach for the screening of known SNPs in M. tuberculosis.
Tuberculosis (TB) remains a major public health concern worldwide, with approximately 8 million new cases and over 2 million deaths each year. Genotyping of Mycobacterium tuberculosis is important for a better knowledge of moving and expanding clones and to better define TB control. The main genotype families have been described, including the Beijing, Haarlem, Africa, East-African-Indian (EAI), and Latin American and Mediterranean (LAM) strains (7, 13). The Haarlem genotype is found worldwide and is associated with widespread epidemics (13). Several methods based on the study of transposable and repetitive elements, which appear to be the main source of genomic diversity in M. tuberculosis, have been developed for epidemiological typing: IS6110-based restriction fragment length polymorphism genotyping (4), spacer oligonucleotide typing (spoligotyping) (12), and MIRU VNTR (mycobacterial interspersed repetitive units-variable number tandem repeats) typing (16). The development of novel reliable rapid and specific methods to distinguish M. tuberculosis genotypes is of great interest to further analyze the spread of the major epidemic clades.
Single nucleotide polymorphisms (SNPs) represent another source of genetic diversity that can provide useful markers for evolutionary and epidemiological studies in clinical M. tuberculosis. M. tuberculosis is a species with a relatively low polymorphism rate, probably linked to an evolutionary recent global dissemination in humans (17, 20). Sequence analysis of 26 structural genes or 24 genes coding for proteins that are targets of the host immune system from phylogenetically diverse M. tuberculosis isolates indicated a very low rate of SNPs (17, 20). Synonymous SNPs or SNPs that do not confer selective advantage are particularly useful for phylogenetic studies because they are not subject to selective pressure. SNPs at codon 463 of the katG gene and at codon 95 of the gyrA gene are the basis for the classification of M. tuberculosis isolates into three principal genotypic groups (20). Several other SNPs have been described as variable between different tubercle bacilli subspecies. For example, a SNP in the narGHIJ promoter region appears to be specific for M. tuberculosis (21), whereas an SNP in oxyR is specific to M. bovis (19). To date, no specific SNPs have been described to differentiate the main epidemic groups.
MgtC (Rv1811) is a virulence factor of M. tuberculosis involved in intramacrophage survival and adaptation to magnesium limitation (3). The M. tuberculosis mgtC promoter has been shown to be a strong promoter that is expressed intracellularly (23). MgtC appears to be a remarkable factor since, to our knowledge, it provides the only clear example of an acquired intramacrophage growth factor shared by several unrelated intracellular pathogens that survive in phagosomes: M. tuberculosis, Salmonella enterica serovar Typhimurium, Brucella suis, and Burkholderia cenocepacia (2, 3, 11, 14). Genes encoding MgtC-like proteins are found in a limited number of eubacterial genomes, and analysis of the phylogeny of MgtC-like proteins and mgtC chromosomal regions suggests that mgtC has been acquired by horizontal gene transfer repeatedly throughout bacterial evolution (1). In mycobacteria, MgtC appears to be absent from rapid growers, and genomic sequencing has confirmed that mgtC is absent from M. smegmatis, suggesting that mgtC might have been acquired after the separation between slow and rapid growers. We have proposed that mgtC of M. tuberculosis is encoded by a 15-kb region that might have been acquired by horizontal transfer (1). Because horizontal gene transfer seems extremely rare in M. tuberculosis (10), the study of mgtC polymorphism might be informative with regard to mgtC evolution. In addition, the study of virulence genes polymorphism is of particular interest to determine whether such genes are under selection pressure.
In the present study, we investigated the polymorphism of mgtC in clinical isolates representative of the genetic diversity of M. tuberculosis to explore the molecular evolution of this virulence gene. We identified an SNP in mgtC that appears to be specific for the Haarlem genotype, and we describe a novel simple PCR-based assay to detect SNPs from mycobacterial cultures.
MATERIALS AND METHODS
Strains and DNAs.
For mgtC sequences, chromosomal DNAs were isolated as described previously (24) from 24 M. tuberculosis strains of various geographic origins representative of the genetic diversity (Table 1) . The collection includes 11 strains from Burkina Faso that are genetically diverse according to spoligotyping genotyping method (S. Godreuil, unpublished data). Isolates from Burkina Faso were cultured on Löwenstein-Jensen medium and were identified as belonging to M. tuberculosis complex by DNA strip assay (Genotype MTBC; Hain Lifescience). These strains were genotyped by using the spoligotyping method (12), and the spoligotyping data were compared to the international database spolDB3.0 (S. Godreuil, unpublished data). In addition, six strains were obtained from C. Gutierrez (Institut Pasteur, Paris, France), and seven strains were obtained from F. Portaels (Prince Leopold Institute of Tropical Medicine, Antwerp, Belgium) (Table 1). The collection of M. tuberculosis isolates includes members of the main epidemic groups (Beijing, Haarlem, EAI, LAM, and T1) (7). Strains or DNAs from other members of the M. tuberculosis complex were obtained from F. Portaels and K. Kremer (National Mycobacteria Reference Laboratory, Bilthoven, The Netherlands). For SNP analysis using the “on/off switch assay,” 29 additional M. tuberculosis chromosomal DNAs extracted from a previously described collection (13) were used (a generous gift from K. Kremer) (Table 2).
TABLE 1.
Characteristics of M. tuberculosis complex strains and mgtC polymorphism based on DNA sequences
| Strain | Sourcea | Species | Groupb | Genotype | Country of isolation | mgtC polymorphismc |
|---|---|---|---|---|---|---|
| CIP 0357 | a | M. tuberculosis | 1 | Beijing | Cambodia | wt |
| CIP 1121 | a | M. tuberculosis | 1 | Beijing | France (Reunion) | wt |
| ITM 022488 | b | M. tuberculosis | 1 | Beijing | Bangladesh | wt |
| ITM 981274 | b | M. tuberculosis | 1 | Beijing | Siberia | wt |
| CIP 1008 | a | M. tuberculosis | 1 | EAI | India | wt |
| CIP 0423 | a | M. tuberculosis | 1 | EAI | France | Val GTC 87 ATC Ile |
| ITM 021038 | b | M. tuberculosis | 1 | EAI | Bangladesh | wt |
| ITM 021071 | b | M. tuberculosis | 1 | EAI | Bangladesh | wt |
| CIP 0443 | a | M. tuberculosis | 2 | Haarlem | Algeria | Arg CGC 182 CAC His |
| CIP 0276 | a | M. tuberculosis | 2 | Haarlem | France | Arg CGC 182 CAC His |
| 14 | c | M. tuberculosis | 2 | Haarlem | Burkina Faso | Arg CGC 182 CAC His |
| 45 | c | M. tuberculosis | 2 | Haarlem | Burkina Faso | Arg CGC 182 CAC His |
| 83 | c | M. tuberculosis | 2 | Haarlem | Burkina Faso | Arg CGC 182 CAC His |
| ITM 8613 | b | M. tuberculosis | 2 | Haarlem | The Netherlands | Arg CGC 182 CAC His |
| ITM 020284 | b | M. tuberculosis | 2 | Haarlem | Bangladesh | Arg CGC 182 CAC His |
| ITM 981269 | b | M. tuberculosis | 2 | Haarlem | Siberia | Arg CGC 182 CAC His |
| 2 | c | M. tuberculosis | 2 | LAM10 | Burkina Faso | wt |
| 32 | c | M. tuberculosis | 2 | LAM10 | Burkina Faso | wt |
| 3 | c | M. tuberculosis | 2 | T1 | Burkina Faso | wt |
| 19 | c | M. tuberculosis | 2 | T1 | Burkina Faso | wt |
| 17 | c | M. tuberculosis | 2 | T1 | Burkina Faso | wt |
| 42 | c | M. tuberculosis | 2 | T1 | Burkina Faso | wt |
| 27 | c | M. tuberculosis | 2 | ST200 | Burkina Faso | wt |
| 15 | c | M. tuberculosis | 2 | ST494 | Burkina Faso | wt |
| CDC 1551 | e | M. tuberculosis | 2 | United States | wt | |
| H37Rv | e | M. tuberculosis | 3 | United States | wt | |
| Erdman | f | M. tuberculosis | United States | Arg CGC 182 CAC His | ||
| ITM 010012 | b | M. africanum | Belgium | wt | ||
| ITM 993123 | b | M. africanum | Belgium | wt | ||
| 2 | d | M. bovis BCG | The Netherlands | wt | ||
| 71 | d | M. bovis BCG | Japan | wt | ||
| 76 | d | M. bovis | Argentina | wt | ||
| 25 | d | M. microti | United Kingdom | wt | ||
| 116 | d | “M. canettii” | Somalia | Val GTT 15 GTC Val |
Source: a, C. Gutierrez; b, F. Portaels; c, S. Godreuil; d, K. Kremer; e, published genomes; f, mgtC sequence from Erdman strain was determined in the course of a previous study (3).
Determined in this study by on/off switch assay (except for CDC1551 and H37Rv).
The wild-type (wt) sequence refers to the one found in the reference strain H37Rv.
TABLE 2.
Characteristics of the M. tuberculosis strains that were tested by the on/off switch assay to analyze the mgtC182 polymorphism
| Straina | Group | Genotypeb | Country of isolation | Amplification by on/off switch assay with
|
|
|---|---|---|---|---|---|
| SNP-182A-R | SNP-182G-R | ||||
| 20 | 1 | Beijing | Mongolia | − | + |
| 30 | 1 | Beijing | South Africa | − | + |
| 111 | 1 | Beijing | South Korea | − | + |
| 8 | 2 | Haarlem | Vietnam | + | − |
| 13 | 2 | Haarlem | Sri Lanka | + | − |
| 51 | 2 | Haarlem | The Netherlands | + | − |
| 84 | 2 | Haarlem | Czech Republic | + | − |
| 87 | 2 | Haarlem | United States | + | − |
| 99 | 2 | Haarlem | Italy | + | − |
| 35 | 2 | Africa | Rwanda | − | + |
| 40 | 2 | Africa | Burundi | − | + |
| 72 | 2 | Africa | Central African Republic | − | + |
| 97 | 2 | Africa | Uganda | − | + |
| 15 | 2 | ST127 | Iran | − | + |
| 16 | 2 | ST4 | Canada | − | + |
| 17 | 2 | ST1727 | Greenland | − | + |
| 18 | 2 | ST211 | United States | − | + |
| 26 | 2 | Zimbabwe | − | + | |
| 27 | 2 | T3 | Ethiopia | − | + |
| 38 | 2 | ST1824 | Tahiti | − | + |
| 41 | 2 | LAM3 | Chili | − | + |
| 56 | 2 | T1 | Curacao | − | + |
| 64 | 2 | Honduras | − | + | |
| 89 | 2 | ST589 | Spain | − | + |
| 98 | 2 | LAM1 | Equador | − | + |
| 108 | 2 | T1 | China | − | + |
| 118 | 2 | LAM3 | Honduras | − | + |
| 12 | 3 | ST131 | Tunisia | − | + |
| 96 | 3 | ST737 | The Netherlands | − | + |
M. tuberculosis strains belong to a previously described collection (13).
The Salmonella enterica serovar Typhimurium strain NM14 harbors a nonpolar chromosomal deletion of the mgtC gene (A. B. Blanc-Potard, unpublished data).
Sequencing of the mgtC gene.
Genomic DNAs were used as a template for PCR amplification of the mgtC gene using specific primers located approximately 80 bp upstream and downstream the mgtC gene: mgtC-F (5′-CGCCTAGGCTCAAACTGCTG-3′) and mgtC-R (5′-CAATACCCGGCGGATCTACC-3′). PCR mix (Goldstar Mix; Eurogentec, Seraing, Belgium) was used with 10 ng of chromosomal DNA and each primer at 200 nM in a total volume of 50 μl. The reactions were initiated with a 5-min denaturation at 95°C, and primer extension was then carried out for 40 cycles as follows: denaturation for 30 s at 95°C, annealing for 30 s at 50°C, a 1-min extension at 72°C, and finally a 5-min extension at 72°C. PCR products were visualized by using 1% agarose gel electrophoresis and were purified by using a NucleoSpin extract II kit (Macherey-Nagel, Düren, Germany). The sequencing was performed by MWG-Biotech (Erbersberg, Germany) with mgtC-F primer. Sequences were performed on both strands when a polymorphism was found.
Detection of mgtC182 SNP by on/off switch PCR assay.
To detect mgtC codon 182 (mgtC182) SNP, we set up a PCR-based technique described earlier as a “on/off swith assay” (25). We used Pfu polymerase (Promega, Madison, WI), a high-fidelity DNA polymerase with 3′ exonuclease activity. Phosphorothioate modification renders primers nuclease resistant. The forward primer SNP-F (5′-GCAAACGCTGACTGTCGC-3′) was common to Haarlem and non-Haarlem genotypes. The reverse primers harbor a 3′ end phosphorothioate-modified nucleotide and are specific to the wild-type (SNP-182G-R, 5′-CTCCGGCCGGCCCCGTGCPS-3′) or mutated (SNP-182A-R, 5′-CTCCGGCCGGCCCCGTGTPS-3′) codon at position 182. The reactions included 4 pmol of primers, 200 μM concentrations of each deoxynucleotide triphosphate, 2 μl of 10× PCR buffer, 0.4 U of Pfu polymerase, and 4 ng of chromosomal DNA template in a final volume of 20 μl. The reactions were initiated with a 2-min denaturation at 95°C; primer extension was then carried out for 30 cycles as follows: 30-s denaturation at 95°C, 30-s annealing at 78°C, and 30-s extension at 72°C, with a final 10-min extension at 72°C. PCR products, 560 bp in length, were visualized by agarose gel electrophoresis. The PCR amplification was also carried out on M. tuberculosis cultures. Bacteria were first inactivated 40 min at 80°C and were boiled for 20 min before storage at −80°C. Assays were performed as described above using 0.4 μl of undiluted culture.
We also used the on/off switch PCR assay for the detection of katG463 and gyrA95 SNPs to define the genetic categories of M. tuberculosis isolates (19). For katG463, the forward primer was katG-F (5′-GCCTTGGGCTCCAGCACG-3′) and the two reverse primers were katG-R1, specific to group 1 (5′-CAGCCTTAAGAGCCAGATCCTPS-3′), and katG-R23, specific to groups 2 and 3 (5′-GCCTTAAGAGCCAGATCCGPS-3′). For gyrA95, the forward primer was gyrA-F (5′-ATGACAGACACGACGTTGCC-3′) and the two reverse primers were gyrA-R12, specific to groups 1 and 2 (5′-GGGCCATGCGCACCAGGGPS-3′), and gyrA-R3, specific to group 3 (5′-GGGCCATGCGCACCAGGCPS-3′). In the on/off switch PCR assay, the annealing temperature was 65°C (katG-R1), 60°C (katG-R23), or 69°C (gyrA-R12 and gyrA-R3).
Cloning of M. tuberculosis mgtC gene and site-directed mutagenesis.
The mgtC gene from M. tuberculosis was amplified by PCR with H37Rv chromosomal DNA as a template (a gift from T. Stinear, Institut Pasteur, Paris, France) and mgtCmtbSph-F (5′-ACATGCATGCAAACGCTGACTGTCGCCG-3′) and mgtCmtbEco-R (5′-GGAATTCTCATTCGGCCTGCGCGTG-3′) primers. The PCR fragment was cloned at the SphI and EcoRI sites of plasmid pNM11, a pBR322 plasmid derivative that harbors the serovar Typhimurium mgtC promoter (A.-B. Blanc-Potard, unpublished data). The resulting plasmid, pNM13, expressed the MgtC protein from M. tuberculosis with transcriptional and translational regulation sites of the mgtC gene of serovar Typhimurium. The Haarlem genotype-specific mutation at codon 182 was introduced in pNM13 by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions and the two complementary primers R182H-F (5′-GCGGGGTATACACACGGGGCCG-3′) and R182H-R (5′-CGGCCCCGTGTGTATACCCCGC-3′). The mutated plasmid was called pNM70.
Complementation experiments in serovar Typhimurium.
The serovar Typhimurium NM14 strain (ΔmgtC) was transformed by electroporation with plasmids encoding MgtC from M. tuberculosis, pNM13 (wild type), and pNM70 (mutation R182H). NM14 was also transformed by pNM11 that harbors only the mgtC promoter and was used as negative control. Two phenotypes were tested in complementation experiments: growth in magnesium-deprived medium and intramacrophage survival. Growth in magnesium-deprived medium was performed in NCE medium supplemented with 10 μM MgCl2 as described previously (2), and the optical density at 600 nm was measured after 18 h of growth. The rate of intramacrophage replication at 18 h postinfection was determined in J774 macrophages as described previously (2).
RESULTS AND DISCUSSION
Polymorphism of MgtC in M. tuberculosis clinical isolates.
We determined the sequence of mgtC in 24 M. tuberculosis clinical isolates (Table 1). These strains have diverse geographic origins and are representative of the M. tuberculosis genetic diversity since they include the main epidemic genotypes. We also included other strains from the M. tuberculosis complex (“M. canettii,” M. africanum, M. microti, M. bovis, and M. bovis BCG) (Table 1). PCR amplification of the mgtC gene was obtained in all strains tested. In addition, we have included the mgtC sequences from the two sequenced genomes of M. tuberculosis (H37Rv and CDC1551) and from the Erdman strain, which was used to construct an mgtC mutant in M. tuberculosis (3). As expected, the polymorphism rate is low (Table 1, three polymorphic sites), and we identified only nonsynonymous mutations in M. tuberculosis (at codons 87 and 182). The number of silent nucleotide polymorphisms in M. tuberculosis is known to be strikingly lower than for most other human bacterial pathogens but is usually greater or similar to that of nonsynonymous polymorphisms (8, 20). The low level of synonymous mutations in mgtC sequences is consistent with the hypothesis of a recent acquisition of the gene in the M. tuberculosis complex by horizontal gene transfer. The amino acid changes associated with both polymorphisms (V87I and R182H) are rather conservative, suggesting that they do not modify MgtC function (see below). Interestingly, one polymorphism at codon 182 appears to be specific to strains of group 2 belonging to the Haarlem genotype (eight strains).
Development of a PCR-based assay to distinguish the mgtC182 polymorphism.
To confirm the association of the mgtC182 polymorphism with the Haarlem genotype, we have extended our analysis to 29 additional M. tuberculosis isolates described by Kremer et al. (13). The additional isolates included mainly strains of group 2 belonging or not belonging to the Haarlem genotype. Several methods have been used to detect SNPs in M. tuberculosis isolates, including analysis of PCR fragments by direct sequencing, hybridization with specific probes, restriction fragment length polymorphism, or single-strand conformation polymorphism (5, 20, 22). Single-step methods based on real-time PCR (20), amplification refractory mutation system (6) or hairpin primers (9) have also been used with diverse efficiencies. In the present study, we developed a simple and reliable method suitable to identify known mutations to distinguish between nucleotide G and A at the second position of codon 182.
We have used a technique called the “on/off switch assay” (25) based on the use of 3′ exonuclease proficient polymerase (Pfu polymerase) and specific primers resistant to this 3′ exonuclease activity due to 3′ phosphorothioate modification (Fig. 1A). The method was set up on two strains that have the wild-type or mutant allele at codon 182 (Fig. 1B), and we subsequently verified the validity of the method on mgtC genes that have been sequenced (not shown). The on/off switch assay was then applied to 29 additional chromosomal DNA to determine the nucleotide at codon 182 of mgtC. All data were unambiguous, and we found that all additional Haarlem strains tested had an A at position 182, whereas all non-Haarlem strains, including strains from group 2, had a G at position 182 (Table 2). Therefore, the SNP identified at codon 182 in mgtC appears to be associated with the Haarlem genotype since it was specifically found in 14 Haarlem strains of very different origin of 63 DNAs tested. According to our results, the Erdman reference strain should belong to the Haarlem genotype. The Erdman strain has not been ascribed to any genotype, but its octal spoligocode (15), which is not found in the SpolDB3.0 database, is very close to the ST47 spoligocode that characterizes Haarlem 1 strains.
FIG. 1.
On/off switch PCR-based assay to analyze the mgtC182 polymorphism. (A) Illustration of the on/off switch assay. The technique uses two sets of primers with an identical forward primer (SNP-F) and variable phosphorothioate-modified “SNP-specific” reverse primers. The SNP-specific primers differ only at the 3′-end nucleotide that is complementary to the wild-type sequence (SNP-182G-R) or the mutant sequence (SNP-182A-R). Two PCRs are performed, one with each of the two SNP-specific primers. Amplification occurs only if the 3′ end of the SNP-specific primer matches the sequence of the template. In case of mismatching, the mismatched phosphorothioate-modified reverse primer is trapped within the exo-domain of the Pfu polymerase during proofreading, blocking DNA polymerization. (B) Analysis of the mgtC182 polymorphism by on/off switch assay. Lanes 1 to 4, optimization of conditions for the assay using chromosomal DNA from Haarlem (lanes 1 to 2, CIP 0443) or non-Haarlem (lanes 3 to 4, CIP 0357) strains; lanes 5 to 16, analysis of the mgtC182 polymorphism using cultures of Haarlem (lanes 5 to 6, strain 14; lanes 7 to 8, strain 45) or non-Haarlem (lanes 9 to 10, ITM 990020; lanes 11 to 12 ITM 981277; lanes 13 to 14, ITM 021038, lanes 15 to 16, ITM 993123) strains. The reverse modified primer is either SNP-182A-R (lanes 1, 3, 5, 7, 9, 11, 13, and 15) or SNP-182G-R (lanes 2, 4, 6, 8, 10, 12, 14, and 16).
A polymorphism in the mutator ogt gene has been found to be preferentially associated with Haarlem strains (18). However, to our knowledge, SNPs specifically associated with epidemic families of M. tuberculosis have not been described thus far. The identification of a Haarlem-specific SNP could be useful in epidemiological studies. The on/off switch assay appears therefore to be a simple, rapid, and specific test to distinguish between Haarlem and non-Haarlem strains. The test works on purified chromosomal DNA as well as with heat-killed cultures of M. tuberculosis (Fig. 1B). In addition, we have shown that the assay can be used without ambiguity to determine other SNPs, such as those located at codon 463 of the katG gene (G-to-T mutation) and at codon 95 of the gyrA gene (G-to-C mutation). We have used this technique to determine the group of M. tuberculosis strains listed in Table 1 according to the classification proposed by Sreevatsan et al. (20). Therefore, the on/off switch assay provides a novel approach for the screen of known SNPs in M. tuberculosis.
The Haarlem-associated mutation does not change the MgtC function.
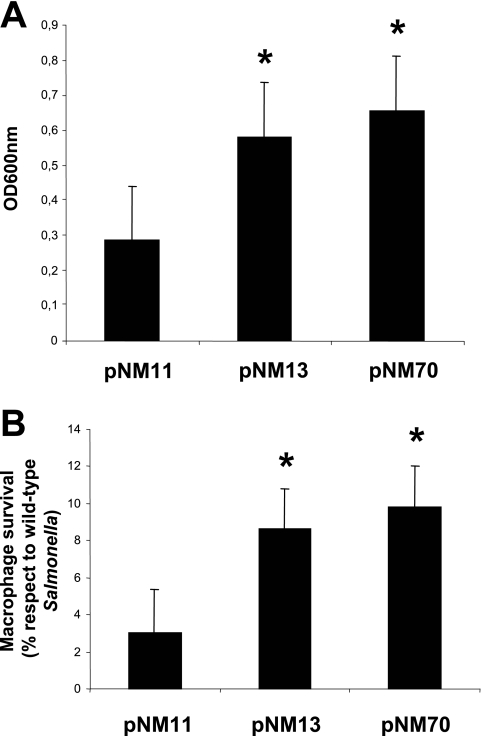
An increased rate of fixation of nonsynomymous mutations relative to synonymous mutations is considered evidence of positive selection acting on improved properties of the corresponding proteins. In addition, the role of MgtC in the virulence of M. tuberculosis has been shown in Erdman strain (3), which harbors the R182H mutation. In this context, we were interested in testing whether the R182H mutation confers a selective advantage to the MgtC protein. To compare the function of M. tuberculosis MgtC proteins harboring an R or H at position 182, we performed complementation experiments in a mgtC mutant of serovar Typhimurium. Heterologous expression of M. tuberculosis MgtC proteins in Salmonella provides a simple and rapid way to test for MgtC-associated phenotypes. The mgtC mutant of S. enterica serovar Typhimurium is defective for growth in low-magnesium medium and replication inside macrophages (2). The introduction of the M. tuberculosis mgtC wild-type gene improves significantly the growth of the Salmonella mutant in low-magnesium medium and its replication in macrophages (Fig. 2). An effect of similar magnitude is found with a MgtC variant found in Haarlem strains that carry a histidine at codon 182 (Fig. 2). Even though we cannot exclude that the behavior of these genes in Salmonella does not exactly reflect their behavior in M. tuberculosis or that there might be slight differences that are not be detected in this assay, this result suggests that the arginine-to-histidine substitution, both of which are polar amino acids, does not have functional impact. The fact that mgtC182 polymorphism does not appear independently on multiple groups of M. tuberculosis strains could also be a feature of a neutral nonsynonymous SNP or could reflect a rare mutation. If the mgtC182 mutation found in Haarlem strains is not driven by adaptative functional modification, it could provide a useful genetic tool to distinguish between Haarlem and non-Haarlem strains.
FIG. 2.
Comparison of the activity of MgtC wild-type and MgtC R182H variant from M. tuberculosis by complementation assay in serovar Typhimurium. Plasmids expressing MgtC wild-type from M. tuberculosis (pNM13) or the MgtC R182H variant (pNM70) were introduced into a serovar Typhimurium ΔmgtC strain. A plasmid without the mgtC gene (pNM11) was used as a negative control. The data are means from three different experiments. Asterisks indicate statistical significance (P < 0.05) compared to pNM11. No significant difference was found between pNM13 and pNM70 plasmids. (A) Complementation assay for growth in magnesium-deprived NCE medium. (B) Complementation assay for replication in J774 macrophages. The percentage of replication comparatively to a wild-type strain of Salmonella serovar Typhimurium is shown.
In conclusion, we propose a novel simple and specific method for the rapid detection of M. tuberculosis Haarlem genotype that provides a useful tool to analyze the spread of this genotype. The specificity of this method has been proved with a limited number of M. tuberculosis isolates that are representative of the genetic diversity that belong to the main genotype families, as well as some minor families, and it will be important to further validate the method with additional isolates, both from major and minor lineages.
Acknowledgments
We thank C. Gutierrez (Institut Pasteur, Paris, France) and F. Portaels (Prince Leopold Institute of Tropical Medicine, Antwerp, Belgium) for providing strains and K. Kremer (National Mycobacteria Reference Laboratory, Bilthoven, The Netherlands) and T. Stinear (Institut Pasteur, Paris, France) for providing DNAs. We thank D. O'Callaghan (Inserm U431, Nîmes, France) for revising the manuscript.
E.A. is supported by INSERM and Région Languedoc-Roussillon. A.-B.B.-P. is supported by the INSERM Avenir program and CANAM.
REFERENCES
- 1.Blanc-Potard, A. B., and B. Lafay. 2003. MgtC as a horizontally acquired virulence factor of intracellular bacterial pathogens: evidence from molecular phylogeny and comparative genomics. J. Mol. Evol. 57:479-486. [DOI] [PubMed] [Google Scholar]
- 2.Blanc-Potard, A. B., and E. A. Groisman. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 16:5376-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchmeier, N., A. Blanc-Potard, S. Ehrt, D. Piddington, L. Riley, and E. A. Groisman. 2000. A parallel intraphagosomal survival strategy shared by Mycobacterium tuberculosis and Salmonella enterica. Mol. Microbiol. 35:1375-1382. [DOI] [PubMed] [Google Scholar]
- 4.Cave, M. D., K. D. Eisenach, P. F. McDermott, J. H. Bates, and J. T. Crawford. 1991. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol. Cell Probes 5:73-80. [DOI] [PubMed] [Google Scholar]
- 5.Dolzani, L., M. Rosato, B. Sartori, E. Banfi, C. Lagatolla, M. Predominato, C. Fabris, E. Tonin, F. Gombac, and C. Monti-Bragadin. 2004. Mycobacterium tuberculosis isolates belonging to katG gyrA group 2 are associated with clustered cases of tuberculosis in Italian patients. J. Med. Microbiol. 53:155-159. [DOI] [PubMed] [Google Scholar]
- 6.Fan, X. Y., Z. Y. Hu, F. H. Xu, Z. Q. Yan, S. Q. Guo, and Z. M. Li. 2003. Rapid detection of rpoB gene mutations in rifampin-resistant Mycobacterium tuberculosis isolates in Shanghai by using the amplification refractory mutation system. J. Clin. Microbiol. 41:993-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filliol, I., J. R. Driscoll, S. D. van, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. A. Dang, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, E. Kassa-Kelembho, M. L. Ho, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. De Waard, C. Sola, and N. Rastogi. 2003. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J. Clin. Microbiol. 41:1963-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, J. W. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazbon, M. H., and D. Alland. 2004. Hairpin primers for simplified single-nucleotide polymorphism analysis of Mycobacterium tuberculosis and other organisms. J. Clin. Microbiol. 42:1236-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsh, A. E., A. G. Tsolaki, K. DeRiemer, M. W. Feldman, and P. M. Small. 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA 101:4871-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunt, T. A., C. Kooi, P. A. Sokol, and M. A. Valvano. 2004. Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infect. Immun. 72:4010-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamerbeek, J., L. Schouls, A. Kolk, A. M. van, S. D. van, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kremer, K., S. D. van, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavigne, J. P., D. O'Callaghan, and A. B. Blanc-Potard. 2005. Requirement of MgtC for Brucella suis intramacrophage growth: a potential mechanism shared by Salmonella enterica and Mycobacterium tuberculosis for adaptation to a low-Mg2+ environment. Infect. Immun. 73:3160-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, Q., C. C. Whalen, J. M. Albert, R. Larkin, L. Zukowski, M. D. Cave, and R. F. Silver. 2002. Differences in rate and variability of intracellular growth of a panel of Mycobacterium tuberculosis clinical isolates within a human monocyte model. Infect. Immun. 70:6489-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musser, J. M., A. Amin, and S. Ramaswamy. 2000. Negligible genetic diversity of Mycobacterium tuberculosis host immune system protein targets: evidence of limited selective pressure. Genetics 155:7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rad, M. E., P. Bifani, C. Martin, K. Kremer, S. Samper, J. Rauzier, B. Kreiswirth, J. Blazquez, M. Jouan, S. D. Van, and B. Gicquel. 2003. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 9:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sreevatsan, S., P. Escalante, X. Pan, D. A. Gillies, S. Siddiqui, C. N. Khalaf, B. N. Kreiswirth, P. Bifani, L. G. Adams, T. Ficht, V. S. Perumaalla, M. D. Cave, J. D. van Embden, and J. M. Musser. 1996. Identification of a polymorphic nucleotide in oxyR specific for Mycobacterium bovis. J. Clin. Microbiol. 34:2007-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stermann, M., A. Bohrssen, C. Diephaus, S. Maass, and F. C. Bange. 2003. Polymorphic nucleotide within the promoter of nitrate reductase (NarGHJI) is specific for Mycobacterium tuberculosis. J. Clin. Microbiol. 41:3252-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Temesgen, Z., K. Satoh, J. R. Uhl, B. C. Kline, and F. R. Cockerill III. 1997. Use of polymerase chain reaction single-strand conformation polymorphism (PCR-SSCP) analysis to detect a point mutation in the catalase-peroxidase gene (katG) of Mycobacterium tuberculosis. Mol. Cell Probes 11:59-63. [DOI] [PubMed] [Google Scholar]
- 23.Triccas, J. A., W. J. Britton, and B. Gicquel. 2001. Isolation of strong expression signals of Mycobacterium tuberculosis. Microbiology 147:1253-1258. [DOI] [PubMed] [Google Scholar]
- 24.Van Soolingen, D., P. E. de Haas, P. W. Hermans, and J. D. van Embden. 1994. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 235:196-205. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, J., K. Li, J. R. Pardinas, S. S. Sommer, and K. T. Yao. 2005. Proofreading genotyping assays mediated by high fidelity exo+ DNA polymerases. Trends Biotechnol. 23:92-96. [DOI] [PubMed] [Google Scholar]