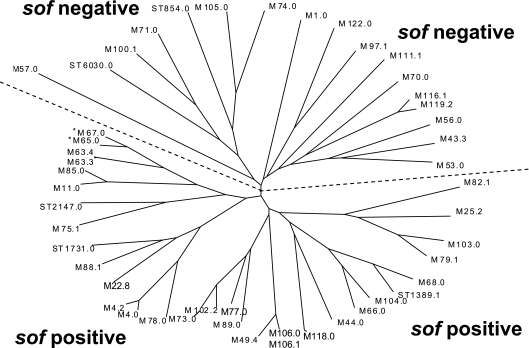
FIG. 1.
Sequence relationships between deduced N-terminal M protein sequences from study set isolates. M protein sequences consisting of 22 signal sequence residues and 66 N-terminal residues were aligned using the Wisconsin Package 10.3 PileUp program. A phylogram was generated as described in Materials and Methods, using the Wisconsin Package version 10.3 Distances and GrowTree programs followed by entering the data into the TreeView program, selecting the unrooted tree option. The asterisks displayed for M65 and M67 indicate the sole exceptions in this study of M sequences from sof-negative and sof-positive strains clustering on the same half of the dendrogram. It is also interesting that one CDC emm65 reference strain apparently contains an aberrant (deletion) sof gene derivative (www.cdc.gov/ncidod/biotech/strep/emmdata.htm#emm65).