Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) infections have become common among both hospitalized and nonhospitalized patients. Optimal outpatient therapy for MRSA infections has yet to be determined, but this matter is complicated by the possibility of inducible macrolide-lincosamide-streptogramin B resistance (MLSBi). We studied the prevalence of MLSBi in community- and hospital-associated S. aureus isolates and the prevalence of community-associated MRSA (CA-MRSA) and identified clinical predictors of CA-MRSA and MLSBi. Among 402 S. aureus isolates, the overall prevalence of MLSBi was 52%, with 50% of MRSA and 60% of methicillin-susceptible S. aureus isolates exhibiting MLSBi. CA-MRSA represented 14% of all isolates and had a lower prevalence of MLSBi than hospital-associated MRSA (33% versus 55%). The presence of skin or soft-tissue infection was predictive for CA-MRSA, and the presence of a comorbidity was predictive for MLSBi. Due to the low prevalence of MLSBi among CA-MRSA isolates, clindamycin remains a useful option for outpatient therapy.
Infections due to methicillin-resistant Staphylococcus aureus (MRSA) have become increasingly common among nonhospitalized patients. Numerous outbreaks of MRSA infections have been recognized in previously healthy people who lacked traditional risk factors for acquisition of MRSA (1, 2, 4-7). Skin and soft-tissue infections (SSTIs) are a common manifestation of staphylococcal disease in many community outbreaks, with invasive staphylococcal disease being less common. Recent work has determined that these community-associated MRSA (CA-MRSA) strains are genetically distinct from health care-associated MRSA (HA-MRSA). Community-associated MRSA strains frequently harbor virulence factors different from those of HA-MRSA strains, particularly the Panton-Valentine leukocidin genes, which are associated with SSTIs (1, 11, 20, 26). In addition, methicillin resistance is conferred by the type IV staphylococcal chromosome cassette mec (SCCmecIV) gene, which is distinct from types I through III, which have previously been found in HA-MRSA strains (17).
As MRSA infections have become increasingly common in the community setting, the development of empirical antimicrobial therapeutic strategies for SSTIs has become more problematic. Clindamycin has long been an option for treating both methicillin-susceptible S. aureus (MSSA) and MRSA infections, particularly for SSTIs. However, expression of inducible resistance to clindamycin could limit the effectiveness of this drug.
Previous reports of inducible macrolide-lincosamide-streptogramin B resistance (MLSBi) among S. aureus isolates were derived from cross-sectional analyses with a wide range of MLSBi prevalences (7% to 94%) (12, 22-25). Data describing MLSBi prevalence or clinical predictors of the presence of MLSBi among CA- and HA-MRSA isolates are quite limited. In the present study, we aimed to characterize MLSBi resistance in both hospital- and community-associated S. aureus isolates, including MRSA and MSSA, at our institution.
(This work was presented in part at the 105th General Meeting of the American Society for Microbiology, 5 to 9 June 2005, Atlanta, GA [M. Patel, D. J. Loebl, Jr., J. L. Morris, M. G. Jones, S. A. Moser, K. B. Waites, and C. J. Hoesley, Abstr. 105th Gen. Meet. Am. Soc. Microbiol., abstr. C-046, 2005.])
MATERIALS AND METHODS
Microorganisms and antimicrobial susceptibility tests.
We prospectively collected sequential nonduplicate S. aureus isolates exhibiting erythromycin resistance and clindamycin susceptibility, as determined by broth microdilution using an automated microbiology instrument (Dade MicroScan, West Sacramento, CA) from April to August 2004. Testing for MLSBi was accomplished by the agar disk diffusion (D test) method in accordance with the recommendations of the Clinical and Laboratory Standards Institute (CLSI) (9).
The anatomic sources of the bacterial isolates and their susceptibilities to other antimicrobials, including oxacillin, gatifloxacin, trimethoprim-sulfamethoxazole, tetracycline, linezolid, and daptomycin, were also recorded. The daptomycin susceptibility test was performed using the agar gradient diffusion (E test) technique in accordance with the manufacturer's instructions (AB Biodisk, Solna, Sweden). All antimicrobial susceptibility tests were interpreted in accordance with the 2004 guidelines published by the CLSI (9).
Medical records for the source patients were reviewed for demographic information, history of prior hospitalization, ZIP code of residence, presence of a major comorbid condition (e.g., diabetes mellitus, renal dysfunction, postsurgical status, malignancy, solid-organ or stem cell transplant, neutropenia, trauma, or burn injury), and antibiotic exposure within the preceding year. Based on available records, determination was made as to whether a clinical infection that was due to the S. aureus isolate cultured was present, as opposed to asymptomatic colonization. Infection was assumed to be present in all cases in which bacterial isolates were derived from blood or cerebrospinal fluid. If no determination could be made based on available records, the presence of infection was considered “unknown.”
MRSA isolates were designated HA-MRSA if the source patient had any of the following risk factors: a history of hospitalization, residence in a long-term care facility (e.g., nursing home), dialysis, or surgery within one year prior to the date of specimen collection; growth of MRSA 48 h or more upon admission to a hospital; presence of a permanent indwelling catheter or percutaneous device at the time of culture; or prior positive MRSA culture prior to this study. If none of the above risk factors were present, the isolate was considered CA-MRSA. This study was approved by the University of Alabama at Birmingham institutional review board.
Statistical methods.
Univariate comparisons utilize the chi-square analysis for categorical results and the Kruskal-Wallis test for continuous variables. The logistic-regression model was used to determine independent factors related to positive D test results for S. aureus isolates (14). The logistic-regression model was also used to determine independent predictors of community-associated S. aureus infection. P values of ≤0.05 were considered significant.
RESULTS
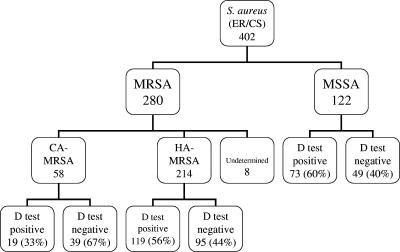
During the study period, 405 S. aureus isolates with the erythromycin-resistant/clindamycin-susceptible phenotype were prospectively collected, of which 402 were available for D test analysis (Fig. 1). Among these, 280 (70%) were MRSA and 122 (30%) were MSSA. The presence of MLSBi was confirmed by the D test for 212 (52%) of the isolates overall, with 139 (50%) MRSA and 73 (60%) MSSA isolates exhibiting MLSBi. Of the 402 staphylococcal isolates, 286 (71%) were classified as health care associated, 99 (25%) were classified as community associated, and 17 (4%) could not be confidently classified and were termed “undetermined.” Positive D test results were noted in 56% and 41% of the health care-associated and community-associated S. aureus isolates, respectively (P = 0.03). One of the undetermined MRSA isolates produced a positive D test result. Among the 280 MRSA isolates, 58 (21%) were designated CA-MRSA, 214 (76%) HA-MRSA, and 8 (3%) undetermined. The presence of MLSBi was detected in 19 (33%) CA-MRSA and 119 (56%) HA-MRSA isolates.
FIG. 1.
Classification of S. aureus isolates. ER, erythromycin resistant; CS, clindamycin susceptible.
The demographic and clinical characteristics of patients with S. aureus infections from whom isolates producing positive versus negative D test results were obtained are outlined in Table 1. The ages, sexes, and races of the patients were not predictive of positive D test results in the univariate analysis. However, isolates with positive D test results were more likely to be health care associated (76% versus 64%, P = 0.03) and associated with patients with any comorbidity (95% versus 81%, P = 0.0001). The presence of infection was slightly higher for isolates with negative D test results (79% versus 70%, P = 0.05). Prior antibiotic exposure showed a strong trend of being predictive of positive D test results (P = 0.08). Multivariate analysis revealed that the presence of any comorbidity in a patient was predictive of a positive D test result (Table 2). Methicillin resistance was not predictive of positive D test results among all S. aureus isolates.
TABLE 1.
Comparison of D-test-positive and D-test-negative S. aureus isolates
| Variable | No. (%) of:
|
P | |
|---|---|---|---|
| Positive D test results (n = 212)a | Negative D test results (n = 189)b | ||
| Isolyte type | |||
| MRSA | 139 (66) | 140 (74) | 0.07 |
| MSSA | 73 (34) | 49 (26) | |
| Classification | |||
| Health care associated | 161 (75) | 120 (64) | 0.03 |
| Community associated | 42 (20) | 57 (30) | |
| Undetermined | 9 (5) | 12 (6) | |
| Sex | |||
| Male | 116 (54) | 102 (54) | 0.921 |
| Female | 96 (46) | 87 (46) | |
| Race | |||
| Caucasian | 137 (64) | 108 (57) | 0.11 |
| African-American | 68 (32) | 77 (41) | |
| Other | 4 (2) | 1 (0.5) | |
| Unknown | 4 (2) | 3 (1.5) | |
| Comorbidity | |||
| Present | 189 (89) | 144 (76) | 0.0001 |
| Absent | 10 (5) | 38 (20) | |
| Unknown | 13 (6) | 7 (4) | |
| Related infection | |||
| Present | 134 (63) | 137 (73) | 0.05 |
| Absent | 57 (26) | 36 (19) | |
| Unknown | 21 (11) | 16 (8) | |
| Prior antibiotic use | |||
| Present | 109 (51) | 81 (44) | 0.08 |
| Absent | 43 (20) | 50 (27) | |
| Unknown | 61 (29) | 58 (29) | |
The mean age of patients in this group was 45 years (range, 0 to 91 years) (P = 0.91).
The mean age of patients in this group was 42 years (range, 0 to 91 years) (P = 0.91).
TABLE 2.
Multivariate analyses
| Isolate group, test result and/or isolate origin, and associated factor | P | OR (95% CI)a |
|---|---|---|
| All S. aureus isolates | ||
| Positive D test results | ||
| Methicillin resistance | 0.017 | 0.55 (0.33-0.90) |
| Comorbidity present | 0.001 | 3.51 (1.56-7.92) |
| Community-acquired status | ||
| Methicillin resistance | <0.0001 | 0.29 (0.17-0.52) |
| Comorbidity present | <0.0001 | 0.09 (0.04-0.22) |
| SSTI | <0.0001 | 7.42 (3.71-14.87) |
| Osteomyelitis | 0.029 | 0.28 (0.09-0.87) |
| MRSA isolates | ||
| Community-associated status | ||
| Comorbidity present | 0.0007 | 0.10 (0.03-0.38) |
| SSTI | <0.0001 | 19.19 (5.89-62.56) |
| Osteomyelitis | 0.081 | 0.17 (0.02-1.24) |
| Prior antibiotic use | <0.0001 | 0.04 (0.01-0.12) |
| Positive D test results and community-associated status | ||
| Comorbidity present | 0.021 | 7.72 (1.37-43.48) |
| SSTI | 0.005 | 0.13 (0.03-0.53) |
CI, confidence interval.
The only positive predictor of community-associated-isolate designation among all S. aureus isolates was the presence of skin and soft-tissue infections (odds ratio [OR] = 7.42; P < 0.0001), while hospital-associated isolates were more likely to be methicillin resistant (OR = 3.45; P < 0.0001) and more likely to be isolated in individuals with comorbidities (OR = 11.11; P < 0.0001) (Table 2). Among patients from whom MRSA isolates were obtained, the community-associated-isolate designation was again linked significantly only to the presence of SSTI (OR = 19.19; P < 0.0001), whereas hospital-associated MRSA isolates were associated with the presence of specific comorbidities (trauma, P = 0.001; recent surgery, P = 0.0003; premature birth, P = 0.01) and prior antibiotic use (P < 0.0001).
Antibiotic susceptibility profiles for MRSA isolates are shown in Table 3. All isolates were erythromycin resistant and clindamycin susceptible (prior to D test analysis). All isolates tested were susceptible to vancomycin, daptomycin, and linezolid. Susceptibilities to trimethoprim-sulfamethoxazole were 100% and 99% among CA- and HA-MRSA isolates, respectively.
TABLE 3.
Antimicrobial susceptibility patterns of MRSA
| Antimicrobial | % of isolates susceptiblea
|
|
|---|---|---|
| CA-MRSA (n = 58) | HA-MRSA (n = 214) | |
| Oxacillin | 0 | 0 |
| Trimethoprim-sulfamethoxazole | 100 | 99 |
| Tetracycline | 89.7 | 92.9 |
| Gatifloxacin | 50 | 42.4 |
| Vancomycin | 100 | 100 |
| Linezolid | 100 | 100 |
| Daptomycin | 100 | 100 |
All isolates were erythromycin resistant and clindamycin susceptible on routine antibiotic testing.
DISCUSSION
Macrolide resistance may be constitutive or inducible in the presence of either a macrolide or a lincosamide inducer. Among MLSBi strains, an inducer promotes production of methylase by erm genes and subsequent methylation of the 23S ribosome, thus making the strain fully resistant to the lincosamides (e.g., clindamycin) and group B streptogramins. Phenotypically, these MLSBi strains appear to be resistant to erythromycin and susceptible to clindamycin on routine antibiotic susceptibility testing. However, inducible resistance can be expressed during a double-disk diffusion test (D test), in which an erythromycin disk will induce clindamycin resistance, thus exhibiting constitutive resistance. Clinically, bacterial strains exhibiting MLSBi have a high rate of spontaneous mutation to constitutive resistance, which could be selected for by use of clindamycin.
Empirical outpatient treatment options for staphylococcal infections have become more limited as concerns about the prevalence of MRSA have increased. In some settings, particularly in outbreaks, the majority of cases of SSTIs are due to MRSA (1, 2, 4, 6, 7). Clindamycin has long been an attractive option because of its efficacy against MSSA and MRSA, its good bone and tissue penetration, and its potential antitoxin effects. Our study suggests that MLSBi is common in both health care-associated and community-associated S. aureus isolates (56% and 33%, respectively). In addition, a significant proportion of the S. aureus isolates at our hospital were designated community acquired by clinical definition (25%), a trend that has been seen nationally (13). In 2003, 72% (897 of 1,246) of all S. aureus isolates collected in the clinical laboratory at the University of Alabama at Birmingham Hospital exhibited the erythromycin-resistant/clindamycin-susceptible phenotype.
Among CA-MRSA isolates, the presence of any of the comorbidities diabetes mellitus, renal dysfunction, postsurgical status, malignancy, solid-organ or stem cell transplant, neutropenia, trauma, or burn injury was a positive predictor of the presence of MLSBi, while presentation with SSTI would suggest the absence of MLSBi. CA-MRSA isolates causing SSTI are likely to be clones of MRSA that have been recognized to be associated with cutaneous abscesses. Specifically, these clones likely have type IV SCCmecA genes.
Inducible MLSB resistance is conferred by the presence of any of the erm A, B, and C genes. A transposon, Tn554, harbors ermA and is generally absent from type IV SCCmecA clones of CA-MRSA, probably explaining the low prevalence of MLSBi among the CA-MRSA isolates in this study (18). The prevalence of MLSBi among CA-MRSA isolates in a pediatric population has been noted to decrease over time, probably representing an expansion of MRSA clones that lack MLSBi genes (8). It is possible that over time, Tn554 or other MLSBi-associated transposons could become part of the genome of CA-MRSA via transduction. We did not identify the SCCmecA gene type in this study but expect that many of our CA-MRSA isolates would be type IV and thus lack the erm genes. CA-MRSA isolates with type IV SCCmecA likely express erythromycin resistance (without lincosamide resistance) through the presence of an efflux pump encoded by the msr(A) gene (3). The implication of this finding is that patients who lack comorbidities and have SSTI due to CA-MRSA may reasonably be offered clindamycin as a treatment option because of a lower likelihood that the strains contain MLSBi. However, among HA-MRSA isolates, the prevalence of MLSBi was significant and should be considered when determining treatment options.
There have been only a few reports describing patients who received clindamycin for S. aureus infections with MLSBi (10, 12, 15, 19, 21, 25). Clinical failures were present in 9 of the 12 (75%) patients described, with development of constitutive MLSB resistance in all but 1 patient. No clinical trials investigating the relationship between MLSBi and clindamycin failures have been performed to date. Currently, some authors recommend avoidance of clindamycin for treatment of complicated infections which may have a high bacterial burden, such as abscesses or osteomyelitis (16). If clindamycin is used for treatment of a less severe MLSBi S. aureus infection, the patient should be closely monitored for signs of failure or relapse of infection. Non-MLSBi infections can be treated with clindamycin if appropriate. The results of this study represent S. aureus isolates from a single hospital and geographic area; the prevalences of MLSBi, CA-MRSA, and specific clones of MRSA may differ in different regions. Clinical microbiology laboratories should adopt testing for MLSBi if local prevalence is found to be substantial, with isolates which exhibit MLSBi reported as being clindamycin resistant.
Acknowledgments
This work was supported in part by grants from Cubist Pharmaceuticals and Pfizer Pharmaceuticals.
We acknowledge the assistance of Donald Loebl and Jason Morris in retrieving clinical information and the technical assistance of Donna M. Crabb, Lynn Duffy, Caroline Reynolds, and Marga G. Jones.
Conflicts of interest are as follows. Mukesh Patel is a member of the Speakers Bureau for Pfizer Pharmaceuticals and Wyeth Pharmaceuticals. Ken B. Waites has received grant support from Pfizer. Craig J. Hoesley is a member of the Speakers Bureau for Cubist Pharmaceuticals.
REFERENCES
- 1.Baggett, H. C., T. W. Hennessy, R. L. Leman, C. Hamlin, D. Bruden, A. Reasonover, P. Martinez, and J. C. Butler. 2003. An outbreak of community-onset methicillin-resistant Staphylococcus aureus skin infections in southwestern Alaska. Infect. Control Hosp. Epidemiol. 24:397-402. [DOI] [PubMed] [Google Scholar]
- 2.Begier, E. M., K. Frenette, N. L. Barrett, P. Mshar, S. Petit, D. J. Boxrud, K. Watkins-Colwell, S. Wheeler, E. A. Cebelinski, A. Glennen, D. Nguyen, and J. L. Hadler. 2004. A high-morbidity outbreak of methicillin-resistant Staphylococcus aureus among players on a college football team, facilitated by cosmetic body shaving and turf burns. Clin. Infect. Dis. 39:1446-1453. [DOI] [PubMed] [Google Scholar]
- 3.Buckingham, S. C., L. K. McDougal, L. D. Cathey, K. Comeaux, A. S. Craig, S. K. Fridkin, and F. C. Tenover. 2004. Emergence of community-associated methicillin-resistant Staphylococcus aureus at a Memphis, Tennessee children's hospital. Pediatr. Infect. Dis. J. 23:619-624. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, K. M., A. F. Vaughn, K. L. Russell, B. Smith, D. L. Jimenez, C. P. Barrozo, J. R. Minarcik, N. F. Crum, and M. A. K. Ryan. 2004. Risk factors for community-associated methicillin-resistant Staphylococcus aureus infections in an outbreak of disease among military trainees in San Diego, California, in 2002. J. Clin. Microbiol. 42:4050-4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. JAMA 282:1123-1125. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2003. Methicillin-resistant Staphylococcus aureus infections among competitive sports participants—Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000-2003. Morb. Mortal. Wkly. Rep. 52:793-795. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2003. Outbreaks of community-associated methicillin-resistant Staphylococcus aureus skin infections—Los Angeles County, California, 2002-2003. Morb. Mortal. Wkly. Rep. 52:88. [PubMed] [Google Scholar]
- 8.Chavez-Bueno, S., B. Bozdogan, K. Katz, K. L. Bowlware, N. Cushion, D. Cavuoti, N. Ahmad, G. H. McCracken, and P. C. Appelbaum. 2005. Inducible clindamycin resistance and molecular epidemiologic trends of pediatric community-acquired methicillin-resistant Staphylococcus aureus in Dallas, Texas. Antimicrob. Agents Chemother. 49:2283-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute/NCCLS. 2004. Methods for antimicrobial susceptibility testing for bacteria that grow aerobically. CLSI/NCCLS M100-S14. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 10.Drinkovic, D., E. R. Fuller, K. P. Shore, D. J. Holland, and R. Ellis-Pegler. 2001. Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J. Antimicrob. Chemother. 48:315-316. [DOI] [PubMed] [Google Scholar]
- 11.Dufour, P., Y. Gillet, M. Bes, G. Lina, F. Vandenesch, D. Floret, J. Etienne, and H. Richet. 2002. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces Panton-Valentine leukocidin. Clin. Infect. Dis. 35:819-824. [DOI] [PubMed] [Google Scholar]
- 12.Frank, A. L., J. F. Marcinak, P. D. Mahgat, J. T. Tjhio, S. Kelkar, P. C. Schreckenberger, and J. P. Quinn. 2002. Clindamycin treatment of methicillin-resistant Staphylococcus aureus infections in children. Pediatr. Infect. Dis. J. 21:530-534. [DOI] [PubMed] [Google Scholar]
- 13.Fridkin, S. K., J. C. Hageman, M. Morrison, L. T. Sanza, K. Como-Sabetti, J. A. Jernigan, K. Harriman, L. H. Harrison, R. Lynfield, and M. M. Farley. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352:1436-1444. [DOI] [PubMed] [Google Scholar]
- 14.Halperin, M., W. C. Blackwelder, and J. I. Verter. 1971. Estimation of the multivariate risk function; a comparison of the discriminant function and maximum likelihood approaches. J. Chronic Dis. 24:125-158. [DOI] [PubMed] [Google Scholar]
- 15.Levin, T. P., B. Suh, P. Axelrod, A. L. Truant, and T. Fekete. 2005. Potential clindamycin resistance in clindamycin-susceptible, erythromycin-resistant Staphylococcus aureus: report of a clinical failure. Antimicrob. Agents Chemother. 49:1222-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis, J. S., and J. H. Jorgensen. 2005. Inducible clindamycin resistance in staphylococci: should clinicians and microbiologists be concerned? Clin. Infect. Dis. 40:280-285. [DOI] [PubMed] [Google Scholar]
- 17.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDougal, L. K., C. D. Steward, G. E. Kilgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGehee, R. F., F. F. Barrett, and F. Finland. 1968. Resistance of Staphylococcus aureus to lincomycin, clindamycin, and erythromycin. Antimicrob. Agents Chemother. 1968:392-397. [PubMed] [Google Scholar]
- 20.Naimi, T. S., K. H. LeDell, K. Como-Sabetti, S. M. Borchardt, D. J. Boxrud, J. Etienne, S. K. Johnson, F. Vandenesch, S. Fridkin, C. O'Boyle, R. N. Danila, and R. Lynfield. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 290:2976-2984. [DOI] [PubMed] [Google Scholar]
- 21.Rao, G. G. 2000. Should clindamycin be used in treatment of patients with infections caused by erythromycin-resistant staphylococci? J. Antimicrob. Chemother. 45:715. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez, M. L., K. K. Flint, and R. N. Jones. 1993. Occurrence of macrolide-lincosamide-streptogramin resistances among staphylococcal clinical isolates at a university medical center: is false susceptibility to new macrolides and clindamycin a contemporary clinical and in vitro testing problem? Diagn. Microbiol. Infect. Dis. 16:205-213. [DOI] [PubMed] [Google Scholar]
- 23.Sattler, C. A., E. O. Mason, and S. L. Kaplan. 2002. Prospective comparison of risk factors and demographic and clinical characteristics of community-acquired, methicillin-resistant versus methicillin-susceptible Staphylococcus aureus infection in children. Pediatr. Infect. Dis. J. 21:910-916. [DOI] [PubMed] [Google Scholar]
- 24.Schreckenberger, P. C., E. Ilendo, and K. L. Ristow. 2004. Incidence of constitutive and inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci in a community and a tertiary care hospital. J. Clin. Microbiol. 42:2777-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siberry, G. K., T. Tekle, K. Carroll, and J. Dick. 2003. Failure of clindamycin treatment of methicillin-resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin. Infect. Dis. 37:1257-1260. [DOI] [PubMed] [Google Scholar]
- 26.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]