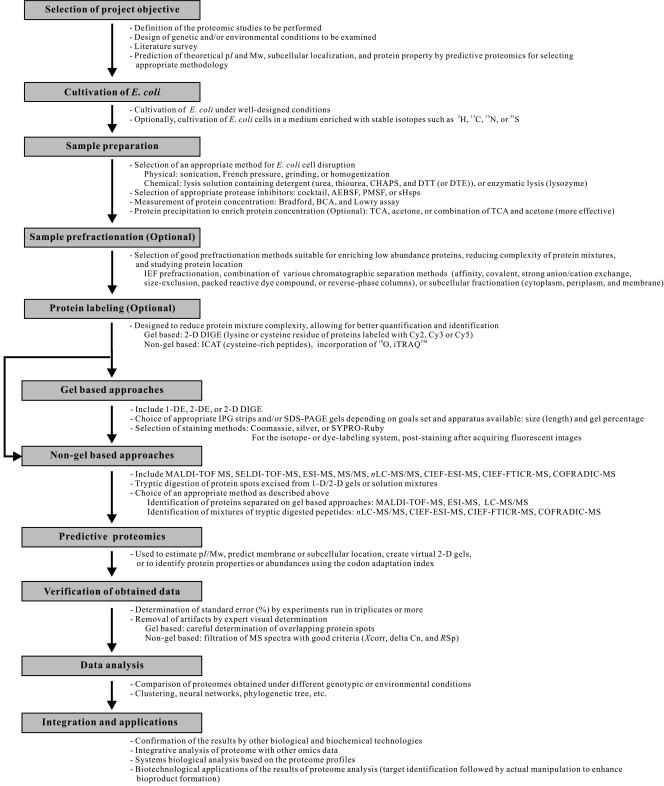
FIG. 2.
General steps for proteomic analysis and tips for success. Once the project objective is set, E. coli cells are cultured and sampled for proteome profiling. During this process, protein samples can be prefractionated or labeled differentially for better comparison of the results. Proteome profiles can be obtained by gel-based and/or non-gel-based approaches. Also, predictive proteomic studies can be performed to analyze a priori the characteristics of proteins in the proteome. Gel-based approaches and non-gel-based approaches are complementary and should be combined if possible to maximize the total number of proteins detected and identified. sHsps IbpA and IbpB were from E. coli and Hsp26 was from Saccharomyces cerevisiae (96). SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; AEBSF, aminoethyl benzylsufonyl fluoride or Pefabloc SC; BCA, bicinchoninic acid; delta Cn, correlation value (difference between the first hit and the second hit); DTE, dithioerythritol; DTT, dithiothreitol; iTRAQ, a multiplexed set of isobaric reagents that yield amine-derivatized peptides (iTRAQ reagents; Applied Biosystems, CA) (253); PMSF, phenylmethylsulfonyl fluoride; RSp, rank preliminary score; SELDI-TOF-MS, surface-enhanced laser desorption ionization-time of flight mass spectrometry; TCA, trichloroacetic acid; Xcorr, cross-correlation (measures how close the spectrum fits to the ideal spectrum).