Abstract
Dipeptide synthesis by aminopeptidase from Streptomyces septatus TH-2 (SSAP) was demonstrated using free amino acid as an acyl donor and aminoacyl methyl ester as an acyl acceptor in 98% methanol (MeOH). SSAP retained its activity after more than 100 h in 98% MeOH, and in the case of phenylalanyl-phenylalanine methyl ester synthesis, the enzyme reaction reached equilibrium when more than 50% of the free phenylalanine was converted to the product. In an investigation of the specificity of SSAP toward acyl donors and acyl acceptors, SSAP showed a broad specificity toward various free amino acids and aminoacyl methyl esters. Furthermore, we applied SSAP to the synthesis of several biologically active peptides, such as aspartyl-phenylalanine, alanyl-tyrosine, and valyl-tyrosine methyl esters.
Some dipeptides exhibit biological activity; for example, aspartyl-phenylalanine methyl ester (AspPhe-OMe) is a high-intensity sweetener, tyrosyl-arginine (TyrArg) is an opioid dipeptide (31), and valyl-tyrosine (ValTyr) inhibits an angiotensin I-converting enzyme's activity (35). In addition, some amino acids, such as tyrosine (Tyr), which is considered as an essential amino acid for some premature infants, have a low solubility. Because these immiscible amino acids become soluble by ligation to another amino acid, alanyl-tyrosine (AlaTyr) is a potential source of tyrosine (10, 11).
Several metallopeptidases, such as thermolysin, synthesize peptides in organic solvents (6, 20, 24). Thermolysin prefers chemically N-protected peptides as substrates for hydrolysis, and therefore N-protected amino acids are used as acyl donors in dipeptide synthesis by thermolysin. Hence, deprotection is required to obtain a biologically active dipeptide. In addition, chemically N-protected amino acids are more expensive than free amino acids and their esters. Thus, from an economical point of view, N-protected amino acids are undesirable for use in the chemoenzymatic synthesis of biologically active dipeptides. Recently, Yokozeki and Hara have reported an efficient enzymatic method involving the aminolysis of ester bonds using a free amino acid and aminoacyl-OMe (38). This method is more cost-effective than the method using thermolysin and has the advantage of using an aqueous solution. Besides the method of Yokozeki and Hara, numerous methods using other enzymes, such as aminoacyl-tRNA synthetase (19), d-alanine ligase (26), and nonribosomal peptide synthetases (14), have been developed. However, these enzymes are too expensive to be applied in the food and pharmaceutical industries. In addition, some biologically active dipeptides, such as TyrArg, AlaTyr, and ValTyr, cannot be synthesized by any of these methods. Thus, the development of a novel method for enzymatic peptide synthesis is crucial for the application of these biologically active dipeptides in the food and pharmaceutical industries.
Recently, we have identified a thermostable aminopeptidase (AP) that contains cocatalytic metallo-active sites and is secreted by Streptomyces septatus TH-2 (SSAP) and succeeded in overproducing it by using recombinant Escherichia coli (1). SSAP has a broad specificity toward peptides (2); however, this enzyme cannot hydrolyze N-protected peptides. Thus, we postulate that SSAP synthesizes a wide variety of dipeptides by using non-N-protected amino acids if SSAP has a function of reverse reaction. In this study, we first demonstrated that SSAP can synthesize dipeptides by using a free amino acid (acyl donor) and aminoacyl-OMe (acyl acceptor) in methanol (MeOH). Next, we investigated its specificity toward the acyl donor and acyl acceptor. Last, we demonstrated that SSAP can be applied to the synthesis of several biologically active peptides.
MATERIALS AND METHODS
Materials.
Free amino acids were purchased from Nacalai Tesque. Aminoacyl-OMes and Leu-p-nitroanilide (pNA) were purchased from Sigma Chemicals. A crude preparation of recombinant SSAP was obtained as follows. E. coli BL21/DE3 harboring pET-SSAP:His, the expression vector for SSAP production (4), was cultivated at 37°C for 12 h in 3 ml of LB medium containing 50 μg/ml kanamycin. The cells were then inoculated into 100 ml of synthetic medium containing 0.5% KH2PO4, 0.5% K2HPO4, 0.44% Na2HPO4, 0.3% (NH4)2SO4, 0.5% glucose, 0.3% MgSO4 · 7H2O, 0.004% FeSO4 · 7H2O, 0.004% CaCl2, 0.00029% CoCl2 · 6H2O, 0.0003% CuSO4 · 5H2O, 0.000036% Na2MoO · 2H2O, 0.001% H3BO3, 0.001% ZnSO4 · 7H2O, 0.2% glycerol, and 50 μg/ml kanamycin (22) and cultivated at 22°C for 12 h. Isopropyl-β-d-thiogalactopyranoside was then added at a final concentration of 0.5 mM, and cultivation was continued for another 24 h under the same conditions. The culture was centrifuged to remove the cells, and the resultant supernatant was used as a crude enzyme preparation.
Preparation of purified SSAP.
Although a His6 tag sequence was fused to the C terminus of recombinant SSAP, it could not be purified by nickel-chelate chromatography. Thus, it was purified as follows. The crude enzyme preparation was brought to 80% saturation with ammonium sulfate. The precipitate obtained by centrifugation was dissolved in 10 mM Tris-HCl containing 1 mM CaCl2 (pH 8.0). It was then heated at 60°C for 30 min with occasional stirring. After centrifugation to remove the precipitate, the solution was dialyzed against 20 mM potassium phosphate buffer (pH 7.0). The dialysate was passed through a hydroxyapatite column (Bio-Rad) equilibrated with the same buffer. The fractions were pooled and dialyzed against 10 mM Tris-HCl (pH 8.0). The dialysate was then loaded onto a Vivapure-Q spin column (Millipore) equilibrated with the same buffer. The bound protein was eluted with 0.2 M NaCl in 10 mM Tris-HCl (pH 8.0). The eluates were pooled and dialyzed against 10 mM Tris-HCl (pH 8.0). The dialysate was used as a purified enzyme preparation.
Enzyme assay.
For routine assay, SSAP activity was determined by continuous spectrophotometric assay with Leu-pNA as the substrate. In the assay, 0.1 ml of 32 mM Leu-pNA was added to 0.9 ml of a mixture containing 100 mM Tris-HCl (pH 8.0) and the enzyme at 37°C. The increase in absorbance at 405 nm due to the release of p-nitroaniline per minute was monitored using a U2800 spectrophotometer (Hitachi). Initial hydrolytic activity was determined from the linear portion of the optical density profile (ɛ405 = 10,600 M−1 cm−1) (27).
Dipeptide synthesis by SSAP.
Dipeptide synthesis by SSAP was performed as follows. Twenty microliters of SSAP solution (1 mg/ml) was suspended in 10 μl of 1 M Tris-HCl (pH 8.0) containing 200 mM amino acid. This suspension was placed in a 1.5-ml microtube, frozen by immersion in liquid nitrogen, and lyophilized with a FreeZone Freeze-Dry Systems model 7679520 (Labconco) at a vacuum of 0.3 hPa for 2 h. This lyophilized enzyme was used for dipeptide synthesis. The activity remained at 78% after lyophilization.
The reaction was initiated by adding 100 μl of 98% MeOH containing 50 mM aminoacyl-OMe to the lyophilized enzyme. The reaction was then continued with vigorous shaking (300 rpm) at 25°C for an appropriate time (20 min to 50 h). Because SSAP could not synthesize dipeptides in >99.8% MeOH solution, the reaction was terminated by adding 1.9 ml of 100% MeOH. After centrifugation, the reaction mixture was analyzed by thin-layer chromatography (TLC) or high-performance liquid chromatography (HPLC).
TLC.
One of the assay methods for dipeptide synthesis was TLC. Three microliters of the reaction mixture was applied to a TLC plate (60 F254; Merck) and developed with n-butanol-acetate-water (7:2:1, vol/vol) for approximately 1 h. Dipeptide, aminoacyl-OMe, and free amino acid were detected by spraying with 2.5% ninhydrin solution dissolved in acetone-water (1:1, vol/vol).
HPLC.
Dipeptides synthesized by SSAP were quantitated with an HPLC system (2690 Separation Module; Waters) equipped with a C18 reverse-phase column (Hydrosphere C18; YMC). The reaction mixture was filtered, and 10 μl of each sample was subjected to chromatography. Each sample was eluted with a solvent A-solvent B gradient of 75:25 to 0:100 for 15 min (1 ml/min), where solvent A was 20 mM KH2PO4 and solvent B was 100% MeOH, and detected by measuring absorbances at 210 and 280 nm with a 490E Programmable Multiwavelength Detector (Waters). The data were processed with the Millennium 32 computer program (Waters Chromatographic Division).
MS.
The molecular mass of each dipeptide synthesized by SSAP was determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) with an Autoflex II TOF/TOF (Bruker Daltonics). The dipeptides synthesized by SSAP were separated and desalted by HPLC under the conditions described in the subsection on HPLC, in which solvent A was substituted with Milli Q water. The separated dipeptides were then dried with a rotary evaporator (centrifugal concentrator model CC105; Tomy) for 2 h. We requested Bruker Daltonics to determine the molecular masses of the lyophilized products. For molecular mass number determination, we chose 2,5-dihydroxybenzoic acid as the MALDI matrix.
Other analytical procedures.
To confirm the composition of the dipeptides obtained, the alkaline and SSAP treatments of dipeptides were performed as follows. The synthesized dipeptides were separated by HPLC and then dried with a rotary evaporator. One hundred microliters of 0.1 M Tris-HCl (pH 8.0) containing 1 μg/ml SSAP or 100 μl of 0.1 M NaOH was added to the lyophilized dipeptides. The dissolved samples were then incubated for 30 min at room temperature. The samples treated with SSAP or alkali were then analyzed by HPLC.
Thermal stability analysis was performed by incubating 200 μl of an enzyme sample (20 μg/ml protein) in 98% MeOH between 4 and 65°C for 30 min. Residual activity was measured under the conditions described in the subsection on the enzyme assay. The effect of the substrate concentration on the reaction rate was examined with a reaction mixture containing 98% MeOH, 0.1 M Tris-HCl (pH 8.0), 200 μg/ml SSAP, 0 to 32 mM free phenylalanine, and 0 to 160 mM Phe-OMe under the conditions described in the subsection on dipeptide synthesis by SSAP. To determine the effect of temperature, the reaction rate of dipeptide synthesis was determined at different temperatures. The stability of SSAP in 98% MeOH was determined by incubating 500 μl of an enzyme sample (20 μg/ml protein) in 98% MeOH at 25°C for an appropriate time. Residual activity was measured under the conditions described in the subsection on the enzyme assay.
RESULTS
Dipeptide synthesis by SSAP.
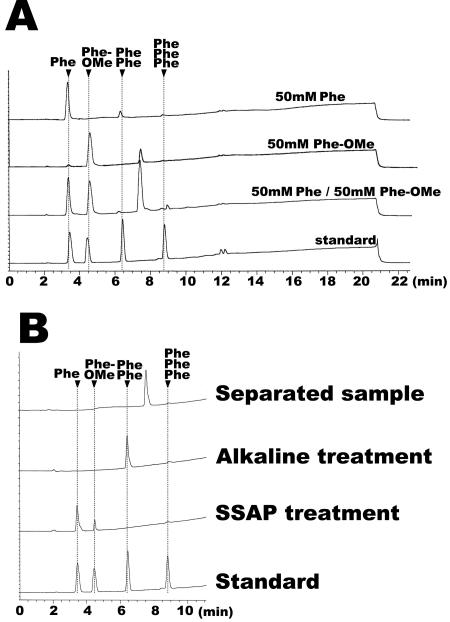
We first selected phenylalanine and its OMe derivative as substrates for demonstrating dipeptide synthesis by SSAP. When only free phenylalanine was used as the substrate, there was a low level of synthesis of PhePhe by SSAP in 98% MeOH for 3 h (Fig. 1A). This indicates that SSAP can synthesize dipeptides in 98% MeOH. When only Phe-OMe was used as the substrate, the HPLC retention time of the product was different from that of PhePhe. It is conceivable that the unknown product was PhePhe-OMe because Phe-OMe was the sole substrate. The level of this product markedly increased with the use of free phenylalanine and Phe-OMe (Fig. 1A).
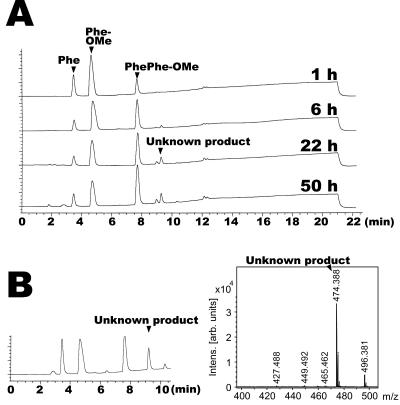
FIG. 1.
HPLC profile of product synthesized by SSAP. (A) HPLC of dipeptide synthesis by SSAP. Free phenylalanine at 50 mM and Phe-OMe at 50 mM were used as substrates. The reaction was performed by using 20 μg of SSAP with vigorous shaking at 25°C for 3 h. (B) HPLC of unknown product. The separated unknown product was treated with alkali or SSAP and then analyzed by HPLC. PhePhe was detected by the HPLC of the product treated with alkali, and free phenylalanine and a small amount of Phe-OMe were detected by treatment of the product with SSAP.
To investigate the structure of the unknown product, we separated the product by HPLC and treated it with SSAP or 0.1 M NaOH. As shown in Fig. 1B, the product treated with alkali was detected at the same retention time as PhePhe. In addition, the components of the product treated with SSAP were free phenylalanine and a small amount of Phe-OMe, indicating that the unknown product was PhePhe-OMe. To corroborate this result, we determined the molecular mass of the unknown product by MALDI-TOF MS. The observed molecular mass of the product was 327 (data not shown), the same as the theoretical molecular mass of PhePhe-OMe.
The MeOH concentration for dipeptide synthesis was investigated. The product PhePhe-OMe was efficiently synthesized in 90 to 98% MeOH. In 99% MeOH, the PhePhe-OMe production was lower than that in 98% MeOH, and PhePhe-OMe synthesis could not be observed in >99.8% MeOH (data not shown). In several organic solvents, including ethanol, isopropanol, and acetone, PhePhe-OMe was effectively synthesized. Of these solvents, MeOH was the most efficient for PhePhe-OMe synthesis (data not shown).
Arrangement of synthesized dipeptide.
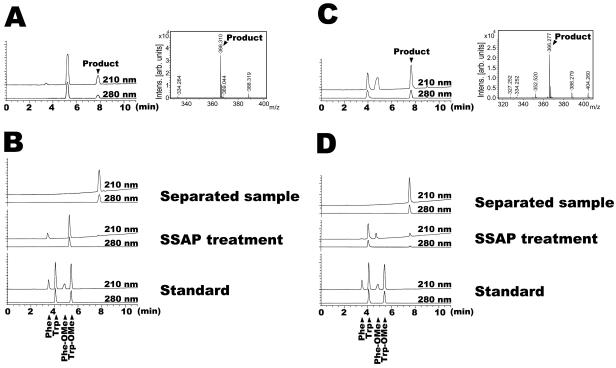
To confirm the arrangement of the product, we chose two model reactions; one was performed with free phenylalanine and Trp-OMe, and the other was performed with free tryptophan and Phe-OMe. The resultant products were then treated with SSAP to confirm the composition. As shown in Fig. 2, the molecular mass of both products was 366 (Fig. 2A and C), indicating that both products were dipeptidyl-OMe composed of phenylalanine and tryptophan. By SSAP treatment, free phenylalanine and Trp-OMe were detected in the product synthesized from free phenylalanine and Trp-OMe (Fig. 2B), indicating that this product was PheTrp-OMe. In contrast, the components of the SSAP-treated product synthesized with free tryptophan and Phe-OMe were free tryptophan and Phe-OMe (Fig. 2D), indicating that this product was TrpPhe-OMe. These results indicate that a free amino acid and aminoacyl-OMe behave as an acyl donor and an acyl acceptor, respectively.
FIG. 2.
Investigation of arrangement of dipeptide synthesized by SSAP. (A and B) HPLC and MS of product synthesized with free phenylalanine and Trp-OMe as substrates (A) and HPLC of this product treated with SSAP (B). The components of this product treated with SSAP were free phenylalanine and Trp-OMe. (C and D) HPLC and MS of product synthesized with free tryptophan and Phe-OMe as substrates (C) and HPLC of this product treated with SSAP (D). Free phenylalanine and Trp-OMe were detected with the treatment of this product with SSAP. In HPLC profiles, the peaks of phenylalanine and Phe-OMe are very small because the absorption coefficient of tryptophan at 210 nm is approximately fourfold higher than that of phenylalanine in this investigation.
Effect of substrate concentration on the rate of dipeptide synthesis by SSAP.
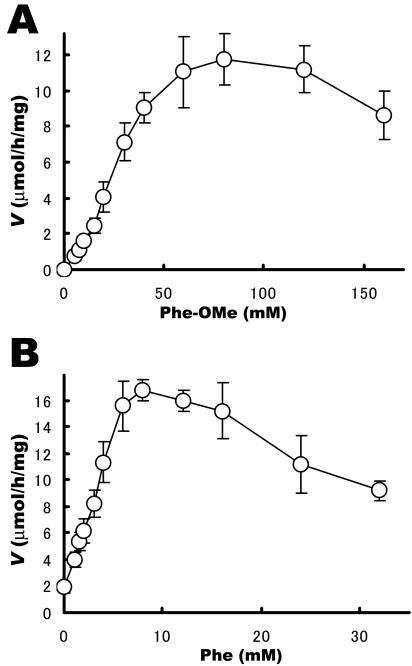
We investigated the effect of the substrate concentration on SSAP activity using free phenylalanine and Phe-OMe. Because we could not obtain the typical nonlinear regression fit to the Michaelis-Menten equation, the kcat and Km for dipeptide synthesis for both substrates could not be calculated. As shown in Fig. 3, the initial velocity is a function of the substrate concentration, with optima at 6 to 12 mM and 60 to 100 mM for free phenylalanine and Phe-OMe, respectively. The anomalous Michaelis-Menten plots in the Fig. 3 are likely due to enzyme inhibition occurring at the highest substrate concentrations. In our previous study, we found that free amino acids have a weak inhibitory effect on the hydrolytic activity of SSAP (Ki, >0.1 M) (1). We speculate that the decrease in the reaction rate of SSAP at free phenylalanine concentrations higher than 12 mM (Fig. 3B) is caused by this inhibitory effect. A similar decrease was also observed in the investigation of the Phe-OMe concentration (Fig. 3B); however, this inhibitory effect of Phe-OMe is weaker than that of free phenylalanine (the decrease in the reaction rate was observed at concentrations higher than 100 mM).
FIG. 3.
Effect of substrate concentration on rate of dipeptide synthesis by SSAP. (A) Effect of acyl acceptor concentration. Free phenylalanine at 50 mM and Phe-OMe at 0 to 160 mM were used as the acyl donor and acyl acceptor, respectively. The reaction was performed by using 20 μg of SSAP with vigorous shaking at 25°C for 2 h. (B) Effect of acyl donor concentration. Free Phe at 0 to 32 mM and Phe-OMe at 50 mM were used as the acyl donor and acyl acceptor, respectively. The reaction was performed by using 20 μg of SSAP with vigorous shaking at 25°C for 2 h. Each value is the average of three independent experiments ± the standard deviation.
Effect of temperature on rate of dipeptide synthesis by SSAP.
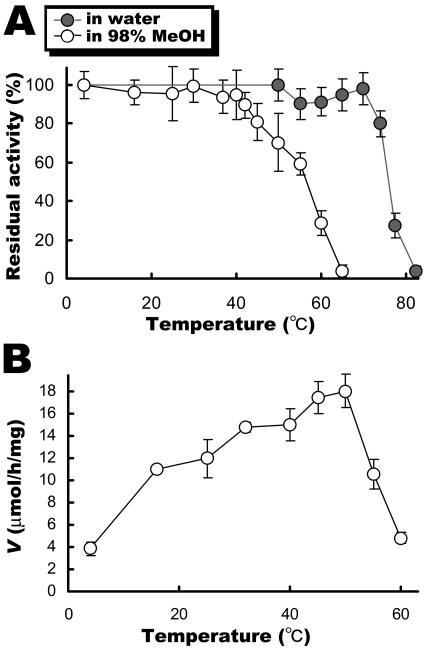
To determine the effect of temperature on SSAP activity, we first examined the thermal stability of SSAP in 98% MeOH. As shown in Fig. 4A, the thermal stability of SSAP in 98% MeOH was lower than that in an aqueous solution. It is believed that this decrease in thermal stability is caused by a decrease in structural stability owing to replacement of water around the enzyme molecule by methanol. The effect of temperature on the rate of dipeptide synthesis by SSAP is shown in Fig. 4B. The activity of SSAP as a function of temperature shows a bell-shaped curve with maximum activity at 50°C.
FIG. 4.
Effects of temperature on stability and rate of dipeptide synthesis by SSAP. (A) Thermal stability of SSAP in aqueous solution and 98% MeOH. Each value is the average of five independent experiments ± the standard deviation. (B) Effect of temperature on dipeptide synthetic activity. Free phenylalanine at 20 mM and Phe-OMe at 50 mM were used as the acyl donor and acyl acceptor, respectively. The reaction was performed by using 20 μg of SSAP with vigorous shaking at an appropriate temperature for 1 h. Each value is the average of three independent experiments ± the standard deviation.
Time dependence of dipeptide synthesis by SSAP.
In dipeptide synthesis by thermolysin, an equilibrium between the substrate and the product was observed (29, 37). Because non-N-protected amino acids are used for dipeptide synthesis by SSAP, synthesized dipeptidyl-OMe may behave as an acyl acceptor. To investigate the reaction equilibrium and the synthesis of undesirable products such as tripeptides, we examined the time dependence of dipeptide synthesis.
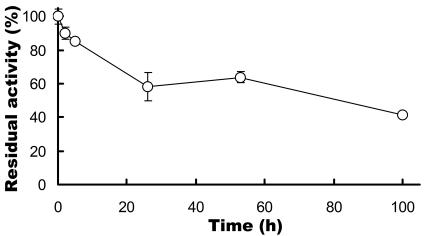
As shown in Fig. 5A, when 20 mM free phenylalanine and 50 mM Phe-OMe were used as substrates, PhePhe-OMe was efficiently synthesized until 6 h, and the quantity of PhePhe-OMe hardly increased after that. In contrast, a small peak was detected (9.3 min in HPLC) after 6 h, and the concentration of this unknown product slightly increased until 50 h. However, there was no increase in the concentration of the unknown product after 50 h (data not shown). As shown in Fig. 6, SSAP retained its activity after 100 h in 98% MeOH, indicating that the reaction equilibrium of dipeptide synthesis by SSAP was reached after 50 h. The rate of free phenylalanine conversion to PhePhe-OMe was 59.8% ± 1.5% when 20 mM free phenylalanine and 50 mM Phe-OMe were used. In contrast to the above results, an increase in the concentration of the unknown product was observed after 22 h with an increase in the quantity of free phenylalanine (50 mM) in the reaction mixture (Fig. 5B). The observed molecular mass of this product was 474, indicating that this product was PhePhePhe-OMe. This result indicates that PhePhe-OMe behaves as a good acyl donor.
FIG. 5.
Investigation of reaction equilibrium. (A) Investigation of reaction equilibrium when 20 mM free phenylalanine and 50 mM Phe-OMe were used as substrates. The reaction was performed by using 20 μg of SSAP with vigorous shaking at 25°C for 1, 6, 22, and 50 h. (B) HPLC profile of product when 50 mM free phenylalanine and 50 mM Phe-OMe were used as substrates for dipeptide synthesis by SSAP. The reaction was performed by using 20 μg of SSAP with vigorous shaking at 25°C for 22 h. Intens., intensity; arb., arbitrary.
FIG. 6.
Stability of SSAP in 98% MeOH. The enzyme sample (20 μg/ml protein) was incubated in 98% MeOH at 25°C for an appropriate time. Residual activity was measured under the conditions described in Materials and Methods. Each value is the average of five independent experiments ± the standard deviation.
Substrate specificity.
In our previous study, we found that SSAP could hydrolyze a wide variety of dipeptides containing hydrophobic residues (2). Thus, we postulate that SSAP can synthesize a wide variety of dipeptides containing hydrophobic residues. To investigate the synthesis of various dipeptides by SSAP, we examined acyl donor specificity by reacting Phe-OMe with 20 different free amino acids for 3 h and acyl acceptor specificity by reacting free phenylalanine with 17 different kinds of aminoacyl-OMe for 3 h.
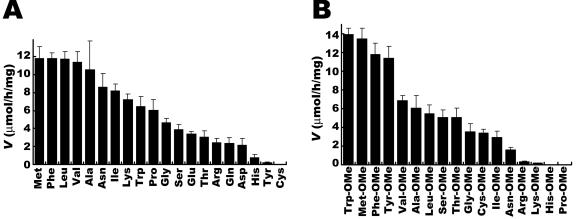
As shown in Fig. 7, a high reaction rate is biased toward synthesis using acyl donors and acceptors that have bulky side chains. In addition, there was no synthesis when free cysteine was used as an acyl donor and His-OMe and Pro-OMe were used as acyl acceptors (Fig. 7). We consider that the nonsynthesis of PhePro-OMe is associated with the fact that SSAP cannot hydrolyze peptides whose penultimate residue is proline (2). Thus, we speculate that SSAP cannot hydrolyze peptides whose N-terminal residue is cysteine and whose penultimate residue is histidine.
FIG. 7.
Substrate specificity of dipeptide synthesis by SSAP. (A) Specificity of SSAP toward acyl donor. (B) Specificity of SSAP toward acyl acceptor. In all cases, 20 mM free amino acid and 50 mM aminoacyl-OMe were used as the acyl donor and acyl acceptor, respectively. The reaction was performed by using 20 μg of SSAP with vigorous shaking at 25°C for 3 h. Each value is the average of three independent experiments ± the standard deviation.
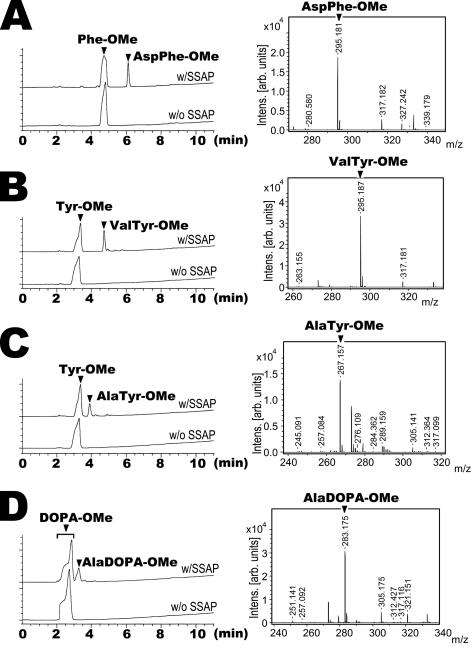
In this investigation, we found that SSAP can synthesize the biologically active peptide AspPhe-OMe, a high-intensity sweetener. Although the rate of AspPhe-OMe synthesis was low, after 24 h, the conversion rate of free aspartate to AspPhe-OMe was 37.1% ± 8.2% (data not shown). The HPLC profile of AspPhe-OMe production and results of the analysis of the molecular mass are shown in Fig. 8A.
FIG. 8.
Synthesis of biologically active dipeptides AspPhe-OMe (A), ValTyr-OMe (B), AlaTyr-OMe (C), and AlaDOPA-OMe (D) by SSAP. In all cases, 20 mM free amino acid and 50 mM aminoacyl-OMe were used as the acyl donor and acyl acceptor, respectively. All panels show the HPLC profiles of the reaction mixture with (w/SSAP) and without (w/o SSAP) the enzyme for the comparison of dipeptide synthesis by SSAP with a negative control. The reaction was performed by using 20 μg of SSAP with vigorous shaking at 25°C for 24 h. (B, C and D) Under the HPLC conditions described in Materials and Methods, the DOPA-OMe and Tyr-OMe peaks formed a shoulder. The hydroxyl groups of the side chain of the substrates may be the cause of the shoulders near these peaks. Intens., intensity; arb., arbitrary.
Synthesis of biologically active dipeptides by SSAP.
We further examined the application of SSAP to the production of several biologically active dipeptides. Because SSAP can synthesize dipeptides whose N-terminal residue is alanine or valine and whose penultimate residue is tyrosine, we tried to synthesize AlaTyr-OMe and ValTyr-OMe. We surmised that other insoluble amino acids, such as 3,4-dihydroxyphenylalanine (DOPA), a therapeutic agent for Parkinson's disease, may increase their solubilities by conversion to dipeptides. Because DOPA has a similar structure to tyrosine, we predict that DOPA-OMe is a good acyl acceptor. Thus, we also tried to convert DOPA into a dipeptide by using free alanine and DOPA-OMe.
As shown in Fig. 8B to D, all of the dipeptides were synthesized by SSAP in 98% MeOH. In these dipeptide syntheses, the conversion rates of free amino acids to dipeptides were above 25% (data not shown). Thus, SSAP is considered applicable to the synthesis of biologically active dipeptides. We further tried to synthesize ester-formed TyrArg by using free tyrosine and Arg-OMe; however, this opioid dipeptide could not be obtained (data not shown). We surmise that tyrosine and Arg-OMe cannot behave as a good acyl donor and a good acyl acceptor, respectively.
DISCUSSION
Bacterial AP was originally studied as a model for understanding the structure and mechanism of action of other peptidases because APs play an important role in many pathological conditions, such as protein maturation, protein degradation, hormone level regulation, and cell cycle control (21, 30, 32, 33, 34). Recent studies demonstrated that bacterial APs are valuable for the preparation of a debittered protein hydrolysate in the food industry (2) and the synthesis of pharmaceuticals (3) because of their broad substrate specificity, high thermal stability, and functions as amidases and esterases (7, 36). In this study, we first demonstrated that one of the bacterial APs, SSAP, has a novel function of dipeptide synthesis in organic solvents.
SSAP belongs to the M28 family, which includes bacterial and human enzymes that accommodate two zinc atoms in their active site. By using APs from Streptomyces griseus and Aeromonas proteolytica as authentic enzymes belonging to the M28 family, the catalytic mechanism of those APs has been extensively studied by structural analysis, inhibitory analysis, and site-directed mutagenesis (4, 8, 9, 12, 13, 15, 16, 17, 18, 28). Thus, the catalytic mechanism of their hydrolytic activity is well known, and SSAP is considered to have the same catalytic mechanism as these enzymes. In this mechanism, a water molecule interacting with an acidic residue is considered to play a role in peptide bond hydrolysis as a nucleophile (17). Because dipeptide synthesis by SSAP occurred in more than 90% MeOH, we speculate that this reaction is due to the deprivation of water molecules, which act as nucleophiles in peptide hydrolysis, caused by the high concentration of MeOH, and the α-amino group of the acyl acceptor behaves as a nucleophile instead of the water molecules under our conditions of dipeptide synthesis by SSAP; however, these remain to be clarified. In addition, an aspect of the mechanism requires further elucidation: SSAP's preference for aminoacyl-OMe as an acyl acceptor.
Compared with known enzymes used for dipeptide synthesis (14, 19, 27, 36, 38), SSAP gave a broad substrate specificity, indicating that it can produce various dipeptides, including several biologically active dipeptides (Fig. 7 and 8). SSAP also shows broad substrate specificities toward peptides and aminoacyl derivatives in terms of hydrolytic activity (1, 2). In particular, SSAP prefers to hydrolyze peptides that have hydrophobic bulky side chains. Similarly, SSAP shows efficient dipeptide synthesis using both acyl donors and acyl acceptors that have bulky side chains. Thus, in dipeptide synthesis by SSAP, peptides that act as good substrates in hydrolysis are appropriate targets of a reverse reaction. This phenomenon is, in effect, the same as the reverse reaction by thermolysin (23, 25).
In this study, we demonstrated that SSAP can be applied to the synthesis of various dipeptides. However, it could not synthesize TyrArg, an opioid dipeptide (31). To perform more convenient dipeptide synthesis, it is crucial to alter the specificities of SSAP toward acyl donors and acyl acceptors. In our recent studies, to make SSAP a convenient biocatalyst for the synthesis of pharmaceuticals, we succeeded in altering its substrate specificity by site-directed mutagenesis (3, 5). In these investigations, we obtained several mutants with high hydrolytic activities toward artificial substrates such as aminoacyl-OMe and aminoacyl amide. However, the hydrolytic activities of these mutants toward peptides are lower than that of wild-type SSAP (3). We previously demonstrated that the specificity toward the N-terminal residue of bacterial APs is affected by the penultimate residue, flanking moiety, and length of peptide substrate (2). From this study, it is suggested that there is a region in bacterial APs associated with the recognition of the penultimate residue of peptide substrates. However, details of this region are unclear. Further study of the recognition of the penultimate residue by SSAP is needed to improve SSAP dipeptide synthesis.
Acknowledgments
This research was supported in part by grants from the Noda Institute for Scientific Research.
REFERENCES
- 1.Arima, J., M. Iwabuchi, and T. Hatanaka. 2004. Gene cloning and overproduction of an aminopeptidase from Streptomyces septatus TH-2, and comparison with a calcium-activated enzyme from Streptomyces griseus. Biochem. Biophys. Res. Commun. 317:531-538. [DOI] [PubMed] [Google Scholar]
- 2.Arima, J., Y. Uesugi, M. Iwabuchi, and T. Hatanaka. 2006. Study on peptide hydrolysis by aminopeptidases from Streptomyces griseus, Streptomyces septatus and Aeromonas proteolytica. Appl. Microb. Biotechnol. 70:541-547. [DOI] [PubMed] [Google Scholar]
- 3.Arima, J., Y. Uesugi, M. Iwabuchi, and T. Hatanaka. 2005. Alteration of leucine aminopeptidase from Streptomyces septatus TH-2 to phenylalanine aminopeptidase by site-directed mutagenesis. Appl. Environ. Microbiol. 71:7229-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arima, J., Y. Uesugi, M. Uraji, S. Yatsushiro, S. Tsuboi, M. Iwabuchi, and T. Hatanaka. 2006. Modulation of Streptomyces leucine aminopeptidase by calcium: identification and functional analysis of key residues in activation and stabilization by calcium. J. Biol. Chem. 281:5885-5894. [DOI] [PubMed] [Google Scholar]
- 5.Arima, J., Y. Uesugi, M. Uraji, M. Iwabuchi, and T. Hatanaka. 2006. The role of Glu196 in environment around substrate binding site of leucine aminopeptidase from Streptomyces griseus. FEBS Lett. 580:912-917. [DOI] [PubMed] [Google Scholar]
- 6.Baek, D. H., J. J. Song, S. J. Kwon, C. Park, C. M. Jung, and M. H. Sung. 2004. Characteristics of a new enantioselective thermostable dipeptidase from Brevibacillus borstelensis BCS-1 and its application to synthesis of a d-amino-acid-containing dipeptide. Appl. Environ. Microbiol. 70:1570-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bienvenue, D. L., R. S. Mathew, D. Ringe, and R. C. Holz. 2002. The aminopeptidase from Aeromonas proteolytica can function as an esterase. J. Biol. Inorg. Chem. 7:129-135. [DOI] [PubMed] [Google Scholar]
- 8.Bzymek, K. P., and R. C. Holz. 2004. The catalytic role of glutamate 151 in the leucine aminopeptidase from Aeromonas proteolytica. J. Biol. Chem. 279:31018-31025. [DOI] [PubMed] [Google Scholar]
- 9.Chevrier, B., C. Schalk, H. D'Orchymont, J. M. Rondeau, D. Moras, and C. Tarnus. 1994. Crystal structure of Aeromonas proteolytica aminopeptidase: a prototypical member of the co-catalytic zinc enzyme family. Structure 2:283-291. [DOI] [PubMed] [Google Scholar]
- 10.Daabees, T. T., and L. D. Stegink. 1978. l-Alanyl-l-tyrosine as a tyrosine source during intravenous nutrition of the rat. J. Nutr. 108:1104-1113. [DOI] [PubMed] [Google Scholar]
- 11.Daabees, T. T., and L. D. Stegink. 1979. l-Alanyl-l-tyrosine as a tyrosine source during total parenteral nutrition. Infusion at 0.5 and 2 mmoles/kg/day in adult rats. Pediatr. Res. 13:894-899. [DOI] [PubMed] [Google Scholar]
- 12.De Paola, C. C., B. Bennett, R. C. Holz, D. Ringe, and G. A. Petsko. 1999. 1-Butaneboronic acid binding to Aeromonas proteolytica aminopeptidase: a case of arrested development. Biochemistry 38:9048-9053. [DOI] [PubMed] [Google Scholar]
- 13.Desmarais, W. T., D. L. Bienvenue, K. P. Bzymek, R. C. Holz, G. A. Petsko, and D. Ringe. 2002. The 1.20 Å resolution crystal structure of the aminopeptidase from Aeromonas proteolytica complexed with tris: a tale of buffer inhibition. Structure 10:1063-1072. [DOI] [PubMed] [Google Scholar]
- 14.Duerfahrt, T., S. Doekel, T. Sonke, P. J. Quaedflieg, and M. A. Marahiel. 2003. Construction of hybrid peptide synthetases for the production of alpha-l-aspartyl-l-phenylalanine, a precursor for the high-intensity sweetener aspartame. Eur. J. Biochem. 270:4555-4563. [DOI] [PubMed] [Google Scholar]
- 15.Fundoiano-Hershcovitz, Y., L. Rabinovitch, Y. Langut, V. Reiland, G. Shoham, and Y. Shoham. 2004. Identification of the catalytic residues in the double-zinc aminopeptidase from Streptomyces griseus. FEBS Lett. 571:192-196. [DOI] [PubMed] [Google Scholar]
- 16.Gilboa, R., H. M. Greenblatt, M. Perach, A. Spungin-Bialik, U. Lessel, G. Wohlfahrt, D. Schomburg, S. Blumberg, and G. Shoham. 2000. Interactions of Streptomyces griseus aminopeptidase with a methionine product analogue: a structural study at 1.53 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 56:551-558. [DOI] [PubMed] [Google Scholar]
- 17.Gilboa, R., A. Spungin-Bialik, G. Wohlfahrt, D. Schomburg, S. Blumberg, and G. Shoham. 2001. Interactions of Streptomyces griseus aminopeptidase with amino acid reaction products and their implications toward a catalytic mechanism. Proteins 44:490-504. [DOI] [PubMed] [Google Scholar]
- 18.Greenblatt, H. M., O. Almog, B. Maras, A. Spungin-Bialik, D. Barra, S. Blumberg, and G. Shoham. 1997. Streptomyces griseus aminopeptidase: X-ray crystallographic structure at 1.75 Å resolution. J. Mol. Biol. 265:620-636. [DOI] [PubMed] [Google Scholar]
- 19.Jakubowski, H. 1995. Synthesis of cysteine-containing dipeptides by aminoacyl-tRNA synthetases. Nucleic Acids Res. 23:4608-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn, D., P. Durrschmidt, J. Mansfeld, and R. Ulbrich-Hofmann. 2002. Boilysin and thermolysin in dipeptide synthesis: a comparative study. Biotechnol. Appl. Biochem. 36:71-76. [DOI] [PubMed] [Google Scholar]
- 21.Lowther, W. T., and B. W. Matthews. 2002. Metalloaminopeptidases: common functional themes in disparate structural surroundings. Chem. Rev. 102:4581-4607. [DOI] [PubMed] [Google Scholar]
- 22.Mishima, N., K. Mizumoto, Y. Iwasaki, H. Nakano, and T. Yamane. 1997. Insertion of stabilizing loci in vectors of T7 RNA polymerase-mediated Escherichia coli expression systems: a case study on the plasmids involving foreign phospholipase D gene. Biotechnol. Prog. 13:864-868. [DOI] [PubMed] [Google Scholar]
- 23.Morihara, K., and H. Tsuzuki. 1970. Thermolysin: kinetic study with oligopeptides. Eur. J. Biochem. 15:374-380. [DOI] [PubMed] [Google Scholar]
- 24.Nakanishi, K., and R. Matsuno. 1990. Continuous peptide synthesis in a water-immiscible organic solvent with an immobilized enzyme. Ann. N. Y. Acad. Sci. 613:652-655. [DOI] [PubMed] [Google Scholar]
- 25.Oka, T., and K. Morihara. 1980. Peptide bond synthesis catalyzed by thermolysin. J. Biochem. 88:807-813. [DOI] [PubMed] [Google Scholar]
- 26.Park, I. S., and C. T. Walsh. 1997. d-Alanyl-d-lactate and d-alanyl-d-alanine synthesis by d-alanyl-d-alanine ligase from vancomycin-resistant Leuconostoc mesenteroides. Effects of a phenylalanine 261 to tyrosine mutation. J. Biol. Chem. 272:9210-9214. [DOI] [PubMed] [Google Scholar]
- 27.Prescott, J. M., and S. H. Wilkes. 1976. Aeromonas aminopeptidase. Methods Enzymol. 45:530-543. [DOI] [PubMed] [Google Scholar]
- 28.Reiland, V., R. Gilboa, A. Spungin-Bialik, D. Schomburg, Y. Shoham, S. Blumberg, and G. Shoham. 2004. Binding of inhibitory aromatic amino acids to Streptomyces griseus aminopeptidase. Acta Crystallogr. D Biol. Crystallogr. 60:1738-1746. [DOI] [PubMed] [Google Scholar]
- 29.Riechmann, L., and V. Kasche. 1986. Reaction mechanism, specificity and pH-dependence of peptide synthesis catalyzed by the metalloproteinase thermolysin. Biochim. Biophys. Acta 8723:269-276. [DOI] [PubMed] [Google Scholar]
- 30.Sanderink, G. J., Y. Artur, and G. Siest. 1988. Human aminopeptidases: a review of the literature. J. Clin. Chem. Clin. Biochem. 26:795-807. [DOI] [PubMed] [Google Scholar]
- 31.Satoh, M., S. Kawajiri, M. Yamamoto, A. Akaike, Y. Ukai, and H. Takagi. 1980. Effects of tyrosyl-arginine (kyotorphin), a new opioid dipeptide, on single neurons in the spinal dorsal horn of rabbits and the nucleus reticularis paragigantocellularis of rats. Neurosci. Lett. 16:319-322. [DOI] [PubMed] [Google Scholar]
- 32.Taylor, A. 1993. Aminopeptidases: structure and function. FASEB J. 7:290-298. [DOI] [PubMed] [Google Scholar]
- 33.Taylor, A. 1993. Aminopeptidase: towards a mechanism of action. Trends Biochem. Sci. 18:167-172. [PubMed] [Google Scholar]
- 34.Taylor, A. 1996. Aminopeptidases, p. 1-20. Landes Bioscience Publishers, Austin, Tex.
- 35.Tokunaga, K. H., C. Yoshida, K. M. Suzuki, H. Maruyama, Y. Futamura, Y. Araki, and S. Mishima. 2004. Antihypertensive effect of peptides from royal jelly in spontaneously hypertensive rats. Biol. Pharm. Bull. 27:189-192. [DOI] [PubMed] [Google Scholar]
- 36.Wagner, F. W., S. H. Wilkes, and J. M. Prescott. 1972. Specificity of Aeromonas aminopeptidase toward amino acid amides and dipeptides. J. Biol. Chem. 247:1208-1210. [PubMed] [Google Scholar]
- 37.Wayne, S. I., and J. S. Fruton. 1983. Thermolysin-catalyzed peptide bond synthesis. Proc. Natl. Acad. Sci. USA 80:3241-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yokozeki, K., and S. Hara. 2005. A novel and efficient enzymatic method for the production of peptides from unprotected starting materials. J. Biotechnol. 115:211-220. [DOI] [PubMed] [Google Scholar]