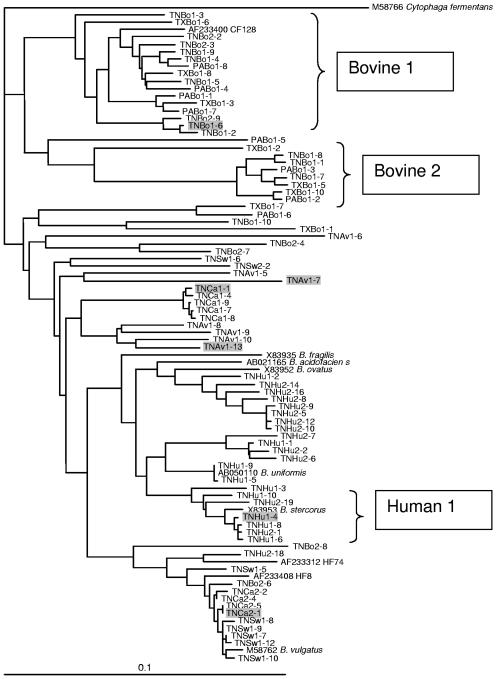
FIG. 1.
Phylogenetic dendrogram showing the relationship of cloned Bacteroides 16S rRNA gene sequences from different animal fecal sources. For each clone in this study, the first two letters represent the state (TN, Tennessee; PA, Pennsylvania; and TX, Texas), the next two letters represent the animal fecal source (Bo, cattle; Eq, horse; Av, chicken; Ca, dog; Sw, swine; and Hu, human), and the final number indicates the individual clone within the library. Sequences were aligned, and a bootstrap consensus tree was created using Clustal X (version 1.64b). The root was determined using the 16S rRNA gene sequence from Cytophaga fermentans (M58766) as an outgroup. References for cultured and uncultured Bacteroides or 16S rRNA gene sequences indicated on the tree were M58766 and M58762 (17); X83935, X83952, and X83953 (32); AB050110 (Y. Miyamoto, unpublished data) and AB021165 (27); and AF233400 and AF233408 (5). Plasmids containing the shaded sequences were used to determine the effect on sequence mismatches on real-time PCR assays in Fig. 2.