TABLE 2.
Technical characteristics of var group primers for SYBR green real-time PCRa
| Characteristic | Value or description for indicated primer pair
|
Value or description for indicated primer pair
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | C1 | C2 | BC1 | BC2 | pvar1utr | pvar2utr | pvar3 | |
| Target region | Grp A DBL1α | Group A exon 2 | Group A exon 2 | upsB | upsB | upsC subtype | upsC subtype | Group B and C exon 2 | Group B and C exon 2 | var1 5′ UTR | var2 5′ UTR | var3 coding |
| Targeted genes in 3D7 predicted by alignments | PFD1235w, MAL7P1.1, PF08_0141, PF11_0008, PFD0020c, PF11_0521, PF13_0003, PFA0015c, MAL6P1.314, PFI1820w | PFD1235w, MAL7P1.1, PF08_0141, PF11_0008, PFD0020c, PF11_0521, PF13_0003, PFA0015c, MAL6P1.314, PFI1820w | PF11_0521, PFD1235w, MAL7P1.1, PF13_0003, PFD0020c, PF11_0008 | MAL6P1.1, PF07_0139, PF08_0142, PF10_0001, PF10_0406, PF11_0007, PF13_0001, PF13_0364, PFA0005w, PFA0765c, PFB0010w, PFB1055c, PFC0005w, PFC1120c, PFD0005w, PFD1245c, PFE0005w, PFI0005w, PFI1830c, PFL0005w, PFL0935c, PFL2665c | MAL6P1.1, PF07_0139, PF08_0142, PF10_0001, PF10_0406, PF11_0007, PF13_0001, PF13_0364, PFA0005w, PFA0765c, PFB0010w, PFB1055c, PFC0005w, PFC1120c, PFD0005w, PFD1245c, PFE0005w, PFI0005w, PFI1830c, PFL0005w, PFL0935c, PFL2665c | PFD0625c, PFL1960w, PF07_0048, PFD0630c, PF07_0049, PF07_0051, PFD0615c, PFD1015c | PFD0615c, PF07_0051, PFD0995c, PFD1000c, PF07_0049, PFD1015c, MAL6P1.252 | PF08_0142, MAL6P1.1, PFA0765c, PFC1120c, PFC0005w, PFE0005w, PF10_0406, PFB1055c, MAL7P1.56, PF08_0140, PF13_0364, PF13_0001, PFL2665c, PFI0005w, PFB0010w, PF11_0007, PF07_0139, PFD1005c, PFD1000c, PF08_0107, PFD0005w, PFL0005w, PFD1015c, PFI1830c, PF07_0050, PF10_0001 | MAL7P1.50, PF07_0048, PF08_0103, PF08_0106, PFA0005w, PFD1245c, PFL0935c, PFL1950w | PFE1640w | PFL0030c | PFA0015c, MAL6P1.314, PFI1820w |
| Coverage in 3D7 (group) | All A genes, including var3 types | All A genes, including var3 types | All A genes, excluding var3 types | All B genes | All B genes | 8/13 C | 7/13 C | 17/22 B, 4/13 C, 1/4 BA, 2/9 BC | 4/9 BC, 3/22 B, 1/13 C | var1 | var2 | var3 |
| Fragment Tm (°C) in 3D7 | 77.2 | 73.6 | 72.1 | 75.4 | 75.6 | 74.1 | 73.5 | 75.9 | 75.5 | 69.6 | 74 | 75.5 |
| Fragment length (bp) | 110-120 | 100 | 160 | 260 | 190 | 106 | 120 | 110 | 170 | 87 | 184 | 155 |
| Fragment Tm (°C)b in field isolates (SD) [no. of PCR-negative genomes/total] | 77.2 (0.4) [0/20] | 73.7 (0.2) [0/20] | 72.2 (0.4) [0/20] | 75.1 (0.4) [0/20] | 75.3 (0.4) [0/20] | 74.2 (0.4) [0/20] | 73.5 (0.3) [0/20] | 75.9 (0.3) [0/20] | 75.6 (0.3) [0/20] | 69.6 (0.2) [0/20] | 74.3 (0.4) [0/20] | 75.6 (0.2) [0/20] |
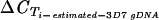 [−log(no. of targeted genes)/log(2)] [−log(no. of targeted genes)/log(2)] |
−3.3 | −3.3 | −2.6 | −4.5 | −4.5 | −3 | −2.8 | −4.7 | −3 | 0 | 0 | −1.6 |
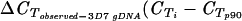 ) ) |
−2.0 | −2.5 | −1.9 | −3.7 | −3.9 | −2.2 | −0.7 | −4.5 | −1.67 | 2.8 | −0.8 | −1.8 |
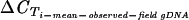 [μ( [μ(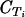 − − 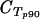 ) (95% CI)]c ) (95% CI)]c
|
−1.2 (0.4) | −1.9 (0.3) | −1.9 (0.3) | −4 (0.2) | −3.6 (0.2) | 0.3 (0.8) | 0.9 (0.5) | −4.55 (0.2) | 0.7 (0.3) | 2.6 (0.2) | 0.3 (0.3) | ND |
The following 3D7 var genes were not targeted by any primer pair: PFD0635c, PF07_0050, PFL1955w, MAL7P1.55, MAL6P1.4, MAL6P1.316, and PFL0020w (three BA and four BC genes).
Mean Tm based on real-time PCR on 20 gDNA and 108 cDNA field isolates.
95% confidence intervals based on real-time PCR on gDNAs from 20 field isolates. The design and validation of primers and probes for the MGB real-time PCR assay were described by Kaestli et al. (16).
