Abstract
We previously reported the enhanced resistance of monoclonal antibodies B6.1 (an immunoglobulin M [IgM]) and C3.1 (an IgG3) against experimental candidiasis. Both MAbs recognize the same fungal epitope. We have since found that a highly passaged B6.1 hybridoma (hp-B6.1) resulted in antibody that has little protective potential. The potential clinical applicability of the antibody and our interest in understanding antibody protection against candidiasis led us to investigate an explanation for this phenomenon. Antibody genetic structure of hp-B6.1, the original hybridoma clone (ori-B6.1) stored frozen since 1995, a subclone of hp-B6.1 that produces protective antibody, the IgG3-producing hybridoma, and a nonprotective IgG1-producing hybridoma were compared. Variable region gene sequences of heavy (VH) and light chains showed genetic instability of VH chains with only the hp-B6.1; the VH sequences from ori-B6.1 and the subclone were, however, identical. Activation-induced cytidine deaminase levels were greatest in the B6.1 hybridomas, which may explain the instability. The constant region CH3 domain remained unchanged, implying normal N-glycation and complement-fixing potential, and antibody binding affinities appeared unchanged. Complement fixation assays surprisingly showed that ori-B6.1 antibody fixes C3 more rapidly than does hp-B6.1 antibody. The VH region primary structure may affect complement activation, which could explain our result. Indeed, antibody from the hp-B6.1 subclone fixed complement like antibody from ori-B6.1. These results show that the greatest protection occurs when antimannan antibodies possess the dual abilities of recognizing the appropriate carbohydrate epitope and rapidly fixing complement; loss of the latter property results in the loss of protective potential by the antibody.
Candida albicans is the most common cause of opportunistic fungal disease in humans (38). The incidence of life-threatening hematogenously disseminated candidiasis, which is predominantly caused by C. albicans, is especially problematic in immunocompromised people ranging from premature infants to AIDS sufferers (1, 15, 49). The choice of anti-Candida drugs is limited, they may adversely affect the host, and the emergence of drug resistance is of potential importance (3, 15, 26, 47). Difficulties often associated with both the diagnosis and treatment (2, 14) support the development of new therapeutic and preventive strategies against candidiasis. The role of antibodies in host defense against fungal diseases is controversial, but it is becoming more widely accepted as the number of publications continue to increase, especially with respect to host defense against cryptococcosis and candidiasis, but with other fungal disease as well (4, 6-9, 12, 16, 30, 32, 35, 45, 46). We are developing vaccines and exploring the efficacies of specific antibodies in aiding the host to resist disseminated candidiasis. Although antibodies have been described that may be directly toxic to this fungus (35, 46), our work has focused on antimannan antibodies, and more information is needed for understanding the basic criteria required for such antibodies to be protective.
During vaccine development, we discovered protective monoclonal antibodies (MAbs) (16, 17, 20-22). The induction of such antibodies through active immunization or passively administered antibodies is predicted to be useful in the prevention and therapy of various forms of candidiasis in both normal and immunocompromised patients. We isolated three isotypes of MAb that recognize the same mannan epitope, β-1,2-mannotriose (18), which is a component of the acid-labile portion of the phosphomannan complex on the cell surface of C. albicans (40, 41). MAbs B6.1 (IgM) and C3.1 (IgG3) are protective against disseminated and vaginal forms of the disease in mouse (16, 21); whereas an IgG1 isotype, MAb G11.2, an apparent derivative of MAb C3.1, is nonprotective. The explanation of the discrepancy in protective activities is likely related to the efficiency by which complement is deposited onto the C. albicans cell surface. We have found that the protective IgM and IgG3 antibodies fix complement very rapidly, whereas a nonprotective IgM (MAb B6) does not. Furthermore, in vivo antibody protection against disseminated candidiasis is complement dependent (19). Mouse IgG1, however, fixes complement very poorly (24, 27, 28). Curiously, monoclonal antibody obtained from the B6.1 hybridoma after successive in vitro passages (highly passaged) showed reduced protective potential, whereas the protective ability of MAb C3.1 remained constant (H. Xin and J. E. Cutler, Abstr. 104th Gen. Meet. Am. Soc. Microbiol. 2004, abstr. H-094, p. 279, 2004). Because of the possible clinical usefulness of antibodies that protect against candidiasis and in an attempt to gain a greater understanding of how antimannan antibodies protect, we pursued an explanation for the loss of protective activity of the highly passaged B6.1 (hp-B6.1).
In this study, the variable region genes of the light (VL) and heavy (VH) chains of each MAb were PCR cloned and sequenced and compared to sequences obtained from the original B6.1 hybridoma (ori-B6.1) which had been stored frozen since 1995 at the American Type Culture Collection (ATCC). The various hybridomas were compared with respect to activation-induced cytidine deaminase (AID) levels, and the antibodies were compared for antigen binding affinities, abilities to fix complements, and protective capabilities. The results indicate an instability potential associated with the B6.1 hybridoma, which may lead to a reduced ability of the antibody to fix complement.
MATERIALS AND METHODS
Organism and culture conditions.
C. albicans CA-1 was used for animal infections and has been previously characterized (16, 20). Cultures for each experiment were started from water stocks and grown as stationary-phase yeast forms in glucose (2%)-yeast extract (0.3%)-peptone (1%) (GYEP) broth under aeration at 37°C. Before use, the yeast forms were collected by centrifugation, washed three times, and suspended in Dulbecco's phosphate-buffered saline (DPBS) to obtain the desired number of yeast cells for intravenous infections of mice as described before (16, 29).
Heat-killed yeast cells from C. albicans strain 3153A were used in the complement fixation assays. This strain (originally obtained from the ATCC; catalog no. 28367) was grown overnight in GYEP at 37°C and centrifuged at 6,000 rpm in a microcentrifuge tube for 2 min, and the pelleted cells were washed five times in sterile deionized H2O. Cells were heat killed at 70°C for 10 min, washed five times as described above, and used immediately or stored for up to 1 month at 4°C.
Mouse hybridoma cells and MAbs.
Hybridoma clones producing MAbs B6.1 (IgM) and C3.1 (IgG3) were generated from mice vaccinated with a phosphomannan complex-liposome preparation as described previously (16, 18). The ori-B6.1 clone (16) was deposited with the ATCC in 1995 and retrieved from that organization in 2004 for the work described here. B6.1 hybridomas put into successive passage since 1995 are called highly passaged B6.1 in this study. Subclones of B6.1 hybridomas were obtained from hp-B6.1 by twice limiting dilution for each.
The hybridoma cell line producing MAb G11.2 (IgG1) was obtained from hybridoma C3.1 as a result of sib selection subcloning methods as described previously (42). MAb G11.2 has the same oligomannoside specificity (β-1,2-linked mannobiose or mannotriose) as MAbs B6.1 and C3.1, as determined by enzyme-linked immunosorbent assay (ELISA) inhibition with synthetic β-linked oligomannosides by methods described previously (33). MAbs B6.1 and C3.1 enhance resistance against experimentally disseminated candidiasis (16, 18, 21) by a complement-dependent opsonization mechanism (19). MAb G11.2 is an IgG1 that does not fix complement very well (24, 27, 28) and does not protect against candidiasis (our unpublished data).
The hybridoma cell lines were initially grown in antibiotic-free RPMI 1640 medium (Sigma) supplemented with 15% fetal bovine serum (GIBCO) and 2 mM l-glutamine (Sigma) at 37°C and in the presence of 5% CO2. For the production of antibodies, the hybridoma clones were grown in antibiotic-free, BD cell MAb serum-free medium (but containing 1.1 mg bovine serum albumin/ml) in a CELLine device (BD, Bedford, MA) and concentrated by using centrifugal filter devices (Centricon Plus-80; Millipore Corporation, Bedford, MA).
RT-PCR and cloning PCR fragments.
Hybridoma RNA was isolated (RNeasy midi RNA isolation kit; QIAGEN) and dissolved in 200 μl RNase-free water and quantified by A260 (total RNA at 100 ng was used for each reverse transcription-PCR [RT-PCR]).
For PCR amplification of mouse MAb variable domains, initial primer constructions were as previously described by Orlandi and coworkers (34); however, the light-chain primer set (VK1BACK/FOR) was selective for the aberrant light-chain sequence coded by the myeloma fusion partner used for production of the hybridomas (5). Therefore, the primer sets for light-chain sequences were changed to those described previously by Clackson and coworkers (10). The light-chain primer VK2BACK (5′-d [GACATTGAGCTCACCCAGTCTCCA]) was used with a mix of four J region primers, MJK1FONX (5′-d [CCGTTTGATTTTCCAGCTTGGTGCC]), MJK2FONX (5′d [CCGTTTTATTTCCAGCTTGGTCCC]), MJK4FONX (5′-d [CCGTTTTATTTCCAACTTTGTCCC]), and MJK5FONX (5′-d [CCGTTTCAGCTCCAGCTTGGTCCC]) to amplify the light-chain kappa families (10). For variable region heavy-chain sequencing, the recommendations of Orlandi et al. (34) were followed. The primer set VH1BACK/FOR (34) for heavy-chain variable regions were VH1BACK (5′-d [AGGTSMARCTGCAGSAGTCWGG] in which S = C or G, M = A or C, R = A or G, and W = A or T) and VH1FOR (5′-d [TGAGGAGACGGTGACCGTGGTCCCTTGGCCCCAG]); these primers were synthesized by Invitrogen. cDNA was synthesized in our lab using a ProSTAR HF single-tube RT-PCR system (high fidelity) (Stratagene). Briefly, a 50-μl reaction mixture contained 100 ng of each RNA, 100 ng of VH1FOR primer or VK2FOR primer, 100 ng of VH1BACK primer or J region primers, 1 μl of dNTP mix (40 mM), and 5.0 μl of 10× HF RT-PCR buffer. Reverse transcriptase StrataScript RT (1.25 U) and TaqPlus Precision DNA polymerase (2.5 U) were added into each reaction mixture after 10 min of incubation at 65°C. Thirty minutes of incubation for the first-strand synthesis was sufficient. After StrataScript RT was inactivated by heat treatment at 95°C for 1 min, a typical cycle was 1 min at 95°C (denature), 1 min at 55°C (anneal), and 2 min at 68°C (extension) for 40 cycles, followed by a 10-min final extension at 68°C.
Sequence analysis was extended to include the CH3 constant region of the heavy chains by use of designed oligonucleotide RT-PCR primers (DG gene program; Accelrys, Inc.). The CH3FWD primer (5′-d [CTTCTTGAAGAACGTGTCTCCAC]) and CH3REV primer (5′-d [GCAGGTACACAGCAGGTGGATG]) were synthesized by Invitrogen.
The PCR products were directly ligated into a pCR2.1 TA cloning vector (TOPO TA cloning; Invitrogen) after removal of excess primers and other contaminants by QIAquick PCR purification (QIAGEN). The ligated constructs were transformed into TOP10 competent cells, and the transfected bacteria were plated onto Luria-Bertani (LB) plates containing 100 μg/ml ampicillin (Sigma) and 40 μg/ml 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) (Sigma). White colonies from transformed plates were screened for inserts by EcoRI digestion (New England) of DNA minipreps from overnight LB-ampicillin cultures. Cultures containing inserts were used for plasmid isolation using the QIAprep miniprep kit (QIAGEN).
DNA sequencing and sequence analysis.
The cloned mouse Ig variable-region inserts were sequenced by use of an ABI Prism 3100 sequencer (Gene Therapy Center, Tulane Health Sciences Center). Sequences were analyzed using the DG gene program (Accelrys, Inc.), and the ClustalW program (EMBL-EMI). All sequence data were confirmed by second-strand sequencing.
Real-time quantitative RT-PCR analysis of AID expression.
Total RNA isolated from hybridomas as described above was of high quality as indicated by an A260/A280 of >1.9 and analysis by agarose gel electrophoresis.
AID PCR primers (5′ primer, 5′-GACAACTAAAGGATGCGCCGAAGTTG [FAM]C-3′; 3′ primer, 5′-CCTGGGAAGGGCTACATGAAA-3′) were designed (LUX Designer software, Invitrogen) and these, along with primers for mouse β-actin (internal control), were synthesized commercially (Invitrogen). The source of the sequence used to construct the AID RT-PCR primers is “Mus musculus activation-induced cytidine deaminase (AID) mRNA, complete cds” (DNA database at EMBL www.ebi.ac.uk/embl; accession no. AF132979). Real-time RT-PCR was performed by using the SuperScript III platinum one-step quantitative RT-PCR system (Invitrogen) in the iCycler iQ detection system. Total RNA (a series of consecutive twofold dilutions) was used in a 50-μl RT-PCR mixture which contained 1 U SuperScript III RT/Platinum Taq mix, 2× SuperScript reaction mix, and 100 nM of each primer. After 15 min of incubation at 50°C and 2 min at 95°C, amplification was performed by 60 cycles at 95°C for 15 s, followed by 60 cycles at 60°C for 30 s. The PCRs were quantified by selecting the amplification cycle where the PCR product of interest was first detected (cycle threshold [CT]). Triplicate experiments were performed for each sample, and CT values were averaged. The relative quantification of AID mRNA expression was normalized to the expression level of the endogenous reference (β-actin) in each sample.
Relative binding affinities of various MAbs.
Relative binding affinities were determined by an inhibition ELISA as described previously (33). In previous studies (18, 33), we showed that the MAbs recognize β-1,2-linked oligomannosides. Of high interest was the finding that binding affinity is inversely related to mannose chain length, except that the antibodies recognize the biose and triose forms of β-1,2-linked oligomannosides with approximately equal affinities. These latter results justified the use of synthetic β-1,2-linked mannotriose in solution as an inhibitor to assess relative binding affinities of the MAbs for immobilized phosphomannan complex antigen isolated from the fungal cell surface. Briefly, C. albicans strain A9 phosphomannan complex obtained by 2-mercaptoethanol extraction of whole cells (18) was dissolved in PBS (10 μg/ml), and the solution was used to coat 96-well ELISA plates (100 μl, overnight at 4°C). The wells were washed five times with PBST (PBS containing Tween 20, 0.05% [vol/vol] and blocked with 2% bovine serum albumin-PBST, 200 μl). MAb B6.1 (IgM) (2.5 mg/ml), C3.1 (IgG3) (1.75 mg/ml) and G11.2 (IgG1) (2.0 mg/ml), produced as concentrated tissue supernatants described above, were diluted to 1:80,000 (B6.1) and 1:3,000 (C3.1 and G11.2) for ELISA measurements. The antibodies were mixed with epitope (inhibitor) (synthetic β-1,2-linked mannotriose, kindly provided by David R. Bundle) dissolved in PBST at a concentration between 0.1 μM and 1 mM, and the resulting solution of each concentration was added to the phosphomannan complex-coated microtiter wells in triplicate and incubated at 21 to 23°C for 3 h. The wells were washed three times with PBST, and goat anti-mouse polyvalent immunoglobulin (IgG, IgA, and IgM) peroxidase-conjugated antibody (diluted 1:10,000 in PBST) (Sigma) 100 μl was added and incubated for 1 h at 21 to 23°C. The wells were washed five times with PBST, followed by the addition of 100 μl of substrate solution (25 ml of 0.05 M phosphate-citrate buffer [pH 5.0], 200 μl of an aqueous solution of O-phenylenediamine 50 mg/ml [Sigma], and 10 μl of 30% H2O2). Color was allowed to develop for 10 min, stopped by the addition of 100 μl of 2 M H2SO4, and read at 492 nm (microtiter plate reader, model 450; Bio-Rad, Richmond, CA). The percent inhibition was calculated relative to wells containing antibody without inhibitor.
As a further test for relative binding affinities, the ELISA inhibition was conducted as described above, but instead of using the mannotriose as the inhibitor, either the intact phosphomannan complex or the acid-labile part containing solubilized beta-linked oligomannosides of various lengths (18) was used as the inhibitor. The test range of inhibitor preparations was between 0.0005 μg/ml and 500 μg/ml.
Passive immunization of mice and assessment of protection.
The MAbs were appropriately diluted in DPBS to give identical agglutination titers against either whole C. albicans CA-1 or phosphomannan complex-coated latex beads (16, 20). Prior to mouse injections, antibody solutions were spun at 15,000 × g for 15 min to remove aggregates. MAbs tested for their ability to enhance resistance against disseminated candidiasis were the original B6.1, B6.1 from the hp-B6.1, and B6.1 from a subclone of the hp-B6.1. The negative control materials tested in mice were MAbs adsorbed with C. albicans yeast cells (16, 20) and DPBS. For each condition, 6- to 8-week-old female C57BL/6 mice (Jackson Laboratories) were given 0.5 ml of the test MAbs or control materials intraperitoneally, followed 4 h later by 0.1 ml intravenously of a suspension containing 5 × 106 yeast cells per milliliter of DPBS. The mice were monitored by a blinded observer for the development of a moribund state, defined as being listless, unable to eat or drink, and nonreactive to probing. At the time that a mouse was deemed moribund, it was sacrificed and that day was recorded and entered along with the statuses of other animals for the subsequent calculation of the median survival time (MST) by use of the Kaplan-Meier statistical analysis (GraphPad Prism, version 4). There were five to six mice per group, and each experiment was repeated four to five independent times. All mouse experiments were performed entirely in our AAALAC-certified animal facility at the Richmond Institute for Children, and all protocols were sanctioned by our Institutional Animal Care and Use Committee.
Complement fixation analysis.
Activation of C3 and binding of C3 fragments to the C. albicans cells were performed in 100-μl complement deposition reaction mixtures consisting of (i) 40% adsorbed fresh normal mouse serum in RPMI 1640 medium with l-glutamine and 25 mM HEPES (Sigma), (ii) 1 μl MAb B6.1 (1.25 μg/μl), and (iii) 5.0 × 106 C. albicans cells. The mixtures were incubated at 37°C and stopped by the addition of 15 μl 100 mM EDTA (Sigma) after 1, 2, 4, 8, and 16 min. Unbound C3 was removed by three washes with ice-cold PBS. The cells were incubated for 1 h at 21 to 23°C in 100 μl of fluorescein (FITC)-conjugated goat IgG fraction to mouse C3 (4 mg/ml; ICN Pharmaceuticals, Inc., Aurora, OH) diluted 1:200 in PBST. The cells were washed three times in DPBS. The yeast cell pellets were suspended in 0.5 ml DPBS and analyzed by flow cytometry (FACSVantage SE; BD Biosciences) equipped with an argon laser excitation at 488 nm. Ten thousand cells in each sample were analyzed (CellQuest Pro software).
To be certain that the above protocol did not yield misleading data due to differential antibody binding kinetics, the following alternative experimental design was also employed. C. albicans cells were incubated with 1 μl MAb B6.1 (1.25 μg/μl) in 60 μl HEPES-buffered RPMI 1640 medium for 30 min at 21 to 23°C, and then 40 μl adsorbed fresh normal mouse serum was added to each reaction mixture. The mixtures were incubated at 37°C and stopped by the addition of 15 μl 100 mM EDTA after 1, 2, and 4 min. The remaining steps were the same as those described above.
Nucleotide sequence accession numbers.
Sequences derived from these studies have been deposited in GenBank (accession numbers DQ273276 to DQ273285).
RESULTS
Highly passaged B6.1 hybridoma is a heterogeneous cell population compared to apparent homogeneity of the C3.1 and G11.2 hybridomas.
In an attempt to understand the loss of protective potential of MAb B6.1, VH and VL sequences from multiple PCR clones were obtained from each of the hp-B6.1, C3.1, and G11.2 hybridomas and compared. A rough approximation of the homo- or heterogeneity of hybridoma cell populations was assessed by sequencing from four to five PCR VL clones and five to nine VH PCR clones of each hybridoma. The VL sequences were 100% identical for all hybridomas (not shown). Mutational changes were noted in VH sequences, however, especially between PCR clones obtained from the hp-B6.1. Whereas five patterns of base changes were noted in the VH sequences of the nine PCR clones from the hp-B6.1 hybridoma, all seven PCR clones were identical from the C3.1 hybridoma and all five PCR clones were identical from the G11.2 hybridoma. In fact, the 12 PCR sequences from the latter two hybridomas were identical except for a single base change at position 21 (Table 1).
TABLE 1.
hp-B6.1 consists of a heterogeneous population of cells distinguished by mutational events at particular locations within the DNA that codes for the variable region domainsa
| Hybridoma/PCR clone | Nucleotide at position in Ig heavy-chain variable region domainb:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FWR1
|
CDR1
|
FWR2
|
CDR2
|
FWR3
|
|||||||||||
| 7 | 9 | 16 | 21 | 103 | 124 | 153 | 166 | 178 | 236 | 237 | 241 | 269 | 271 | 297 | |
| hp-B6.1 | |||||||||||||||
| 1 | A | G | G | T | G | G | A | G | G | C | C | A | T | G | C |
| 2 | A | G | G | A | A | G | A | G | G | C | T | A | T | G | A |
| 3 | A | A | G | T | G | G | A | G | G | C | T | A | T | G | A |
| 4 | C | G | C | T | G | A | A | G | G | C | T | A | T | G | A |
| 5 | C | G | C | A | A | A | A | G | G | C | T | A | T | A | A |
| C3.1 | C | G | G | A | A | G | T | A | A | G | T | G | C | G | C |
| G11.2 | C | G | G | T | A | G | T | A | A | G | T | G | C | G | C |
VH transcripts were PCR amplified and cloned, and several clones derived from each hybridoma were selected for VH sequencing in both directions. Sequence comparisons between clones from the hp-B6.1 were compared to sequences from the C3.1 and G11.2 hybridomas. Possible point mutations from hp-B6.1 are noted by bold type. With the exception of G11.2 at position 21, C3.1 and G11.2 sequences were identical at all positions. Furthermore, the seven cloned PCR-amplified sequences from the C3.1 hybridoma were identical, and the five cloned PCR-amplified sequences from the G11.2 were identical, suggesting that, whereas the hp-B6.1 hybridoma consists of a heterogeneous cell population with respect to VH sequences, the C3.1 and G11.2 hybridomas may each be homogeneous.
Nucleotide positions are based on germ-line Ig alignment.
ori-B6.1 is more homogeneous than the hp-B6.1.
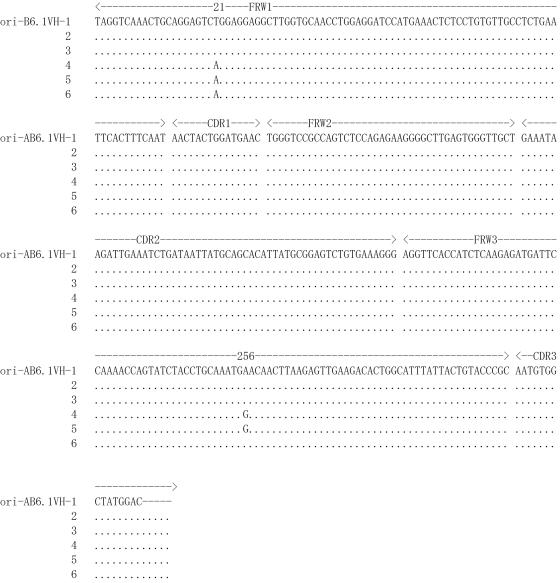
Whereas the multiple hp-B6.1 VH PCR clones showed genetic instability at several positions as indicated in Table 1, sequence consistency was found among six PCR clones obtained from the ori-B6.1 clone that was deposited frozen at the ATCC in 1995. There were only two variations noted in framework regions (FWR): in FWR1, three out of six clones have A instead of T at position 21, and in FWR3, two of six clones have G instead of A at position 256 (Fig. 1).
FIG. 1.
Sequence consistency was found among six PCR clones obtained from the ori-B6.1 clone that was deposited frozen at ATCC in 1995. The Kabat-defined FWR and CDR of the VH gene are indicated. Identities between the compared sequences are shown by dots, and gene mutations are shown by single uppercase letters. The nucleotide alignment of VH obtained from the ori-B6.1 shows only two variations, at positions 21 and 256 in the FWR regions.
The deduced amino acid sequences of the VH segments of the six hp-B6.1 clones indicate several changes within both the FWR and complementarity-determining region 1 (CDR1). The positions of these changes, relative to any of the six clones from ori-B6.1, are shown in Fig. 2A. Relative to the PCR clones from hp-B6.1, the ori-B6.1 showed only a single amino acid shift from asparagine (N) to aspartate (D) at position 83 in FWR3 (Fig. 2B).
FIG. 2.
The amino acid sequences predicted from PCR clones of the B6.1 VH region show greater variability associated with the hp-B6.1 than with the ori-B6.1. (A) The alignment of the deduced amino acid sequences of the VH regions of six consecutive PCR clones derived from the hp-B6.1 (VH-1 to -6) is compared to that of ori-B6.1 (VH-1). Several amino acid changes are predicted, as indicated by the letters shown within both the FWR and CDR1. (B) Alignment of the deduced amino acid sequences of six PCR clones obtained from the ori-B6.1 (VH-1 to -6) showed only a single amino acid shift from asparagines (N) to aspartic acid (D) in clones 5 and 6 at position 83 in FWR3.
Based on the reported mouse VH/VL germ line sequences, the B6.1, C3.1, and G11.2 VH sequences had the highest degree of sequence homology with germ line VH22.1 segments (GenBank accession number, X03398). All VL sequences had the highest homology with germ line IgVκ at four segments (GenBank accession number AJ231225) (data not shown).
Glycosylation sites on the CH3 domains of ori-B6.1 and hp-B6.1 are identical.
We previously reported the relationship between rapid complement fixation and the ability of antibodies to protect (19). We examined the CH3 domain for sequence changes that may affect N-glycosylation, which is critical to the ability of mouse IgM antibodies to fix complement (31). Based on known sequences of mouse CH, CH3-specific primers were designed that resulted in the expected 340-bp PCR product for the CH3 domain of B6.1 (data not shown). Nucleotide sequence alignment of CH3 regions of both ori-B6.1 and hp-B6.1 showed only a single base shift (data not shown) which did not affect the predicted amino acid sequences (data not shown). Therefore, none of the three potential N-glycosylation sites (Asn-X-Ser/Thr) were altered in the CH3 domain of either clone.
AID level is highest in the B6.1 hybridomas.
The exact homology of the VL sequences, along with the high degree of sequence homology of the VH regions, and the specificity of all of the antibodies for the same epitope suggest that the three mAbs were derived from the same germ line. In addition, there are several consistent variations in the CDR and FWR of the VH, which may relate to somatic hypermutation (SHM) and affinity maturation. Along these lines, ∼45% of the mutations are in G or C nucleotides within the so-called hot-spot motifs RGYW (A/G, G, C/T, and A/T) and WRCY that are frequently targeted by AID in SHM both in vivo and in vitro (37, 48), which we addressed below.
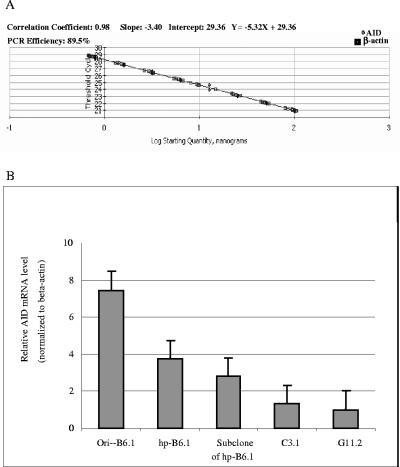
The relative gene expression of AID by ori-B6.1, hp-B6.1, a subclone of hp-B6.1 (VH gene sequence of which is 100% identical with that of ori-B6.1), C3.1, and G11.2 was determined by real-time RT-PCR. The CT value was normalized by the housekeeping gene β-actin, which was amplified at the same efficiency in the same RT-PCR (Fig. 3A). The results showed the highest AID level by the B6.1 hybridomas compared to those of the IgG3 and IgG1-producing hybridomas (Fig. 3B). These findings support that AID activity may be at least in part responsible for the instability of the B6.1 hybridoma. Furthermore, the high activity in the ori-B6.1 suggests to us that AID might induce mutations with continuous in vitro passage of this clone and lead to a heterogeneous cell population of the hp-B6.1 hybridoma. The AID mRNA level in the subclone of hp-B6.1 that maintained the same VH sequence as the ori-B6.1 was lower than that of ori-B6.1, which provides further support that AID is responsible, at least in part, for the observed VH mutations.
FIG. 3.
Relative gene expression of AID showed the greatest expression level in the ori-B6.1, followed by the hp-B6.1. A subclone of hp-B6.1, which has the same VH sequence as that of the ori-B6.1, had intermediate AID activity, whereas the lowest relative AID activity was associated with the highly stable C3.1 and G11.2 hybridomas. (A) Standard curves for β-actin (large squares) and AID (small squares). The CT value was normalized by the housekeeping gene β-actin, which was amplified at the same efficiency in the RT-PCR. (B) Relative gene expression of AID in all the MAb-producing hybridomas was determined by real-time RT-PCR. Note the relatively high AID levels in the B6.1 clones compared to those in C3.1 and G11.2. Error bars indicate standard deviations.
Reduced protection of MAb B6.1 from hp-B6.1 is not due to reduced affinity for the fungal cell wall epitope.
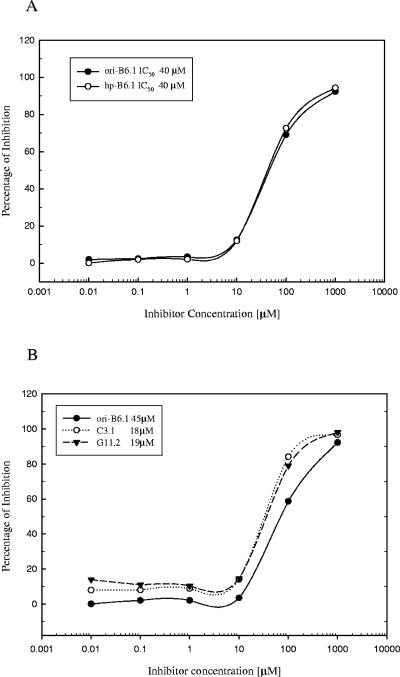
The relative affinities of both IgM B6.1 monoclonal antibodies for the same fungal cell surface epitope were almost identical, as shown in Fig. 4A. In the ELISA inhibition assays, the concentration of epitope (inhibitor) required to achieve 50% inhibitory concentration was less for C3.1 and G11.2 compared to that for MAb from the B6.1 hybridomas (Fig. 4B), indicating the predicted higher affinities of IgG isotypes for the epitope. Because of the haptenic size of the epitope (a trimannose), these data reflect relative binding affinities and not avidity. In addition, however, relative binding affinities were similar when the soluble inhibitor tested was either the intact phosphomannan complex or the acid-labile fraction containing the beta-linked oligomannosides of various lengths (data not shown). These data indicate that the decreased protective activity of MAb B6.1 obtained from the hp-B6.1 is not due to a loss of binding affinity for the fungal cell surface.
FIG. 4.
The relative binding affinities of the various antibodies are similar. Synthetic β-1,2-mannotriose was used as described before (25) to obtain relative binding affinities by an ELISA inhibition assay for MAbs obtained from ori-B6.1, hp-B6.1, C3.1, and G11.2 hybridomas. Each point is the mean of three determinations, and the data shown are from a typical experiment of four independent experiments, all producing similar results. Relative binding affinities were also similar when the soluble inhibitor tested was either the intact phosphomannan complex or the acid-labile fraction containing the beta-linked oligomannosides of various lengths (data not shown). These data suggest that the mutations resulting in amino acid changes in the VH of the hp-B6.1 do not result in binding affinity changes. (A) ELISA inhibition data for the antibodies from ori-B6.1 and hp-B6.1. IC50, 50% inhibitory concentration. (B) ELISA inhibition data for the original ori-B6.1, C3.1, and G11.2 monoclonal antibodies.
MAb from ori-B6.1 is more protective than MAb from hp-B6.1.
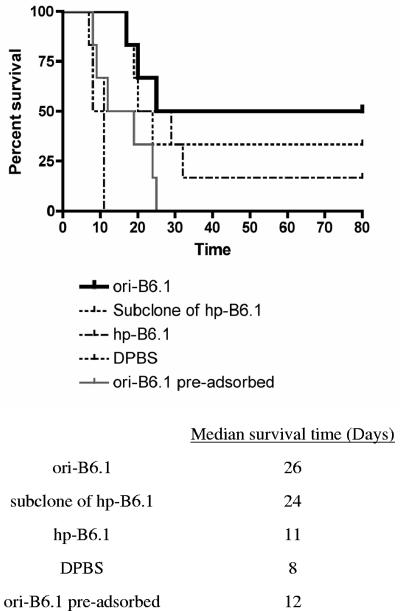
Normal mice were intraperitoneally treated with buffer (DPBS) or standardized doses of MAbs from ori-B6.1, hp-B6.1, a subclone of hp-B6.1, and MAb ori-B6.1 preadsorbed with yeast cells (negative control) prior to an intravenous challenge with 5 × 105 viable C. albicans CA-1 cells. Mice pretreated with MAb from either hp-B6.1 or preadsorbed MAb from ori-B6.1 did not show significant protection relative to animals pretreated with buffer (Fig. 5). However, as expected, MAbs from the ori-B6.1 and from the subclone of hp-B6.1 significantly (P < 0.05) increased the MST of the mice against disseminated candidiasis compared to that of mice given only DPBS buffer before challenge.
FIG. 5.
MAb obtained from ori-B6.1 showed significant protection compared with MAb from hp-B6.1. Normal mice were given the same dose of each of MAbs from ori-B6.1, hp-B6.1, the subclone of hp-B6.1, preadsorbed MAb ori-B6.1 control, and buffer (DPBS) control intraperitoneally. Four hours later, the animals were challenged intravenously with 5 × 105 viable C. albicans cells and susceptibility to disseminated candidiasis was assessed by survival curves. Animals given the MAb from ori-B6.1 had significantly increased median survival times compared with mice that received DPBS (P < 0.05). Very similar results were obtained when MAb from a subclone of hp-B6.1, which has the same VH sequence as the ori-B6.1, was used (P < 0.05). However, mice pretreated with either MAb from hpB6.1 or preadsorbed antibody control did not show significant protection compared with the controls that received DPBS.
MAb B6.1 from ori-B6.1 fixes C3 more rapidly than does MAb B6.1 from hp-B6.1.
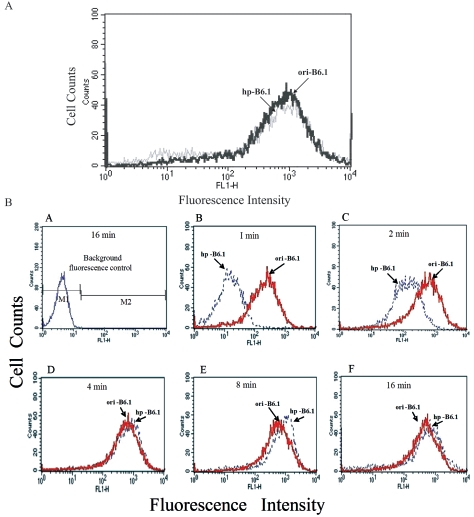
Since the mechanism of protection by MAb B6.1 is dependent on rapid fixation of complement (19), experiments were designed to compare MAbs B6.1 from ori-B6.1 and hp-B6.1 with regard to their ability to rapidly fix C3 to the surface of C. albicans. To be certain that the antibodies were at similar functional concentrations, the chosen concentration of each was adsorbed to the surface of heat-fixed yeast cells, then reacted with anti-μ chain-specific fluoresceinated antibody, and fluorescence was quantified by flow cytometry (Fig. 6A). At the specified dilution of each, fluorescence was essentially identical for both test antibodies. Each antibody was allowed to react with a constant concentration of yeast cells suspended in 40% fresh mouse serum preadsorbed with yeast cells. The reaction times were 1, 2, 4, 8, and 16 min at 37°C; at each time point, the reaction was stopped by the addition of 10 mM EDTA. After reactivity with FITC-goat (IgG) anti-C3 antibody, the amount of C3 binding to yeast cells was determined by flow cytometry. In the absence of either test antibody, yeast cells did not show appreciable fluorescence (Fig. 6B, panel A), which served as the background fluorescence. In the presence of MAb B6.1 from ori-B6.1, fluorescence intensity was markedly greater by 1 min of incubation than the low intensity in the presence of MAb B6.1 from hp-B6.1. This difference was also noted at 2 min of incubation, albeit to a lesser extent (Fig. 6B, panels B and C). By 4 min of incubation, the amount of fluorescence that was detectable on the yeast cells was similar for both test antibodies (Fig. 6B, panels D to F). To rule out the possibility that those differences in complement fixation kinetics were due to differences in the kinetics of antibody-antigen binding, each antibody was also incubated with a constant concentration of yeast cells 30 min prior to the addition of the fresh mouse serum. The complement fixation results were essentially identical at the 1-, 2-, and 4-min time points regardless of the protocol format. These data confirmed that MAb B6.1 from ori-B6.1 fixed C3 to the surface of yeast cells very rapidly relative to MAb B6.1 from hp-B6.1.
FIG. 6.
MAb from ori-B6.1 fixes complement more rapidly than does MAb from hp-B6.1. (A) The standardized concentration of each MAb was adsorbed to the cell surface of heat-fixed C. albicans cells and then reacted with goat anti-IgM/μ-FITC-conjugated antibody for analysis by flow cytometry as described in Materials and Methods. Histograms show the overlay of fluorescence profiles of yeast cells incubated with either of the test antibodies. Note that both antibodies were at similar functional concentrations. (B) The amount of C3 binding to yeast cells in the presence of each test antibody was determined by flow cytometry. The fluorescence intensity is shown on the x axis, and the cell number is shown on the y axis. Histograms show background fluorescence in the absence of either test antibody (A) and the overlay of fluorescence profiles of cells incubated with either MAb from ori-B6.1 or MAb from hp-PB6.1 at 1 min of incubation (B), 2 min of incubation (C), 4 min of incubation (D), 8 min of incubation (E), and 16 min of incubation (F). Note that the ability of MAb from hp-B6.1 to rapidly fix C3 to the fungal cell surface was diminished compared to that of MAb from ori-B6.1, especially within the first 2 min.
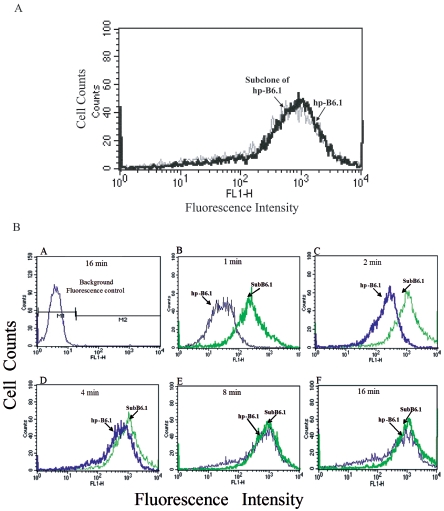
The ability of antibody from the subclone of hp-B6.1, which has the same VH sequence as ori-B6.1, to fix C3 to the fungal cell surface was also assessed and compared to that of MAb from hp-B6.1. The test antibodies were standardized (Fig. 7A). C3 deposition onto yeast cells was obvious by 1 min of incubation in the presence of the antibody from the subclone of hp-B6.1, whereas complement was barely detectable when the antibody from hp-B6.1 was tested (Fig. 7B, panel B). By 4 min, both antibodies complement opsonized the yeast to approximately the same extent (Fig. 7B, panel D).
FIG. 7.
MAb from the subclone of hp-B6.1 restored the ability to rapidly fix complement to the C. albicans cell surface relative to MAb from hp-B6.1. (A) The functional concentration of the test antibodies (MAbs from a subclone of hp B6.1 and hp-B6.1) was standardized, and fluorescence was quantified by flow cytometry. Note that the fluorescence was almost identical for both test antibodies. (B) The ability of antibodies from the subclone of hp-B6.1 and hp-B6.1 was assessed as described before. Similarly, C3 deposition onto yeast cells was obvious by 1 min of incubation in the presence of MAb from the subclone of hp-B6.1, whereas complement was barely detectable when MAb from hp-B6.1 was tested (B). By 4 min, both antibodies complement opsonized the yeast to approximately the same extent (D).
DISCUSSION
Our previous work has indicated that, for antibody to be protective against candidiasis, both specificity and complement-fixing abilities are important (16, 18, 19). Subsequent to those studies, we observed a decline in the ability of one of the protective antibodies, MAb B6.1 (IgM), to passively protect mice; the degree of decline increased as a function of hybridoma passage. Throughout the studies, however, the protective ability of MAb C3.1 (IgG3) remained constant. A third monoclonal antibody, G11.2 (IgG1), was also included in this work as it recognizes the same oligomannoside epitope as MAbs B6.1 and C3.1 do but has never showed protective activity (data not shown). In addition to a fundamental interest in understanding the basis for loss of protective activity of MAb B6.1, these studies are important because caveats about possible antibody instabilities may be critical to the development of a clinically useful product.
The identical epitope specificities, but protectiveness associated with only MAb B6.1 and C3.1, led us initially to compare the VH/VL gene sequences of MAbs B6.1, C3.1, and G11.2. High sequence homology of the V regions from each MAb indicates that they derived from the same germ line gene, which is also supported by a best match with the known VH/VL (Vκ) germ line gene sequences (23, 39).
Although the VL sequences were 100% identical for all hybridomas, the sequence differences in VH domains of MAbs B6.1, C3.1, and G11.2 showed several consistent variations in both CDR and FWR. Importantly, an analysis of the mutational features showed a targeting bias of about 45% occurring in G or C nucleotides within RGYW or WRCY hot spots targeted in SHM by AID both in vivo and in vitro (37, 48). These results suggest that AID is required for SHM in these hybridomas and may have primary responsibility for mutations at these nucleotides. In addition, the DGYW (D = A/G/T) and WRCH (H = T/C/A) motifs reported by Rogozin and Diaz (36) provide a more comprehensive description of the Ig mutation hot spot by AID and we indeed detected three additional mutations by searching these motifs.
The unexpected result was that multiple PCR clones obtained from the hp-B6.1 showed evidence of genetic instability at several locations in both FRW and CDR1 of the VH gene; some of these differences are predicted to result in amino acid changes. More strikingly, sequence consistency was found among six PCR clones obtained from an original B6.1 clone that was deposited frozen at the ATCC in 1995 shortly after discovery of the clone. Therefore, we hypothesized that the apparent propensity of B6.1 to mutate may explain the loss of protective potential by this antibody. To investigate the basis for the genetic instability of hypermutation in hp-B6.1 hybridoma, we examined the expression of the AID gene in mutating and nonmutating hybridoma clones. Quantitative RT-PCR was performed, and VH region mutation rates in different hybridoma clones were found to be roughly correlated with the level of their AID mRNA. Specifically, the mRNA levels of AID for both ori-B6.1 and hp-B6.1 were higher than those for the C3.1 and G11.2 clones, suggesting that AID may explain the mutational propensity of the B6.1 hybridomas. The lower AID level in the subclone of hp-B6.1 that contains no mutation further supports that AID may be the cause for the observed mutations. Furthermore, the relatively low AID levels of the IgG-producing clones also correlate with their stability. The results of these studies should help in the elucidation of genetic and primary structural characters of protective and nonprotective antibodies against candidiasis. The potential significance of this new finding is that hybridomas may be made more stable by the disruption or deletion of the AID gene(s).
To pursue the possible explanation for the decreased protective ability of MAb from hp-B6.1, the MAbs were compared with regard to their abilities to bind the same fungal surface haptenic epitope β-1,2-mannotriose. Almost identical binding activities were found of MAbs from ori-B6.1 and hp-B6.1 for the β-linked mannotriose epitope. In addition, the relative binding affinities of these MAbs were the same when the ELISA inhibition test incorporated either the intact phosphomannan complex or the acid-labile fraction containing the full display of β-linked oligomannosides of varous lengths as the soluble inhibitors (data not shown). We conclude that reduced protection of MAb B6.1 from hp-B6.1 is not due to reduced affinity because the detected amino acid shifts in the VH gene of hp-B6.1 did not affect the antigen binding characteristics of the test antibodies. However, IgG isotypes MAbs C3.1 and G11.2 showed expected higher affinities (33) for the epitope compared to those of the IgM MAbs from the B6.1 hybridomas.
The important role of N-glycosylation in the complement-fixing activities of IgM antibodies led us to examine ori-B6.1 and hp-B6.1 for possible N-glycosylation-site mutations. The CH3 domain of IgM, which corresponds to the Cγ2 domain of the IgG isotype, is the primary locus of interaction with C1q, and mutational changes in this region can affect complement activation (31, 44, 50). CH3 sequence data, however, showed no indication of genetic changes. While this information tends to rule out CH3 primary structural changes as an explanation for the loss of protective potential of MAb from hp-B6.1, the experiments do not exclude the possibility that the hp-B6.1 hybridoma has developed aberrant glycosylation ability.
The protective activity of antibodies against disseminated candidiasis was compared to that of MAb from ori-B6.1 by MST. As expected, MAb from ori-B6.1 showed an ability to provide significant protection when results for animals with this monoclonal antibody (MAb) were compared to results for control animals that received buffer or antibody preadsorbed with yeast, but MAb B6.1 from hp-B6.1 was much less protective when tested at the same concentration. The most likely consideration for loss of protective activity is related to the mutational changes located in the VH region of hp-B6.1 because the MAb from a subclone of hp-B6.1, which has the same VH sequences as that of ori-B6.1, protected mice as well as antibody from ori-B6.1.
By use of a fluorescence assay, the relative abilities of antibodies to fix complement onto the fungal cell surface were compared. In these studies, antibody concentrations were standardized to give the same amount of antibody binding to the cell surface of C. albicans prior to determining the rate of complement activation (C3 deposition on the cell surface). The rate of complement deposition was initially much faster when MAb from ori-B6.1 was tested relative to MAb from hp-B6.1. The rapid rate, 1 to 2 min, may well explain the mechanism of protection. According to our hypothesis, upon entry of yeast into the bloodstream, the events that take place within the ensuing minute or so are critical to determining where in the body C. albicans becomes associated. Rapid complement activation promoted by protective antibody will favor the subsequent association of the fungus with host phagocytic cells, whereas a delay in complement opsonization that occurs with nonprotective antibodies or with alternative complement pathway activation (52) may result in a higher probability of C. albicans becoming associated with nonphagocyte-associated host sites (11).
In our studies, the sequences of the VH regions of hp-B6.1 revealed four amino acid changes encoded by the V genes. Although these changes comprise part of the first hypervariable region for CDR1, which is involved in antigen recognition and may affect epitope affinity (43), the antibodies we studied have the same epitope binding characteristics and similar relative affinities (estimated by the 50% inhibitory concentration), which rules out loss of protective activity due to affinity changes. Additionally, as discussed above, changes in glycosylation sites do not account for differences in C3 activation kinetics.
Our data suggest that changes in the CDR1 and/or changes in FWR1 to -3 of the VH gene may affect C activation by these antibodies. Antibody from the subclone of hp-B6.1, which is protective and has a VH sequence identical to that of ori-B6.1, gave the same result of rapid C3 activation as did antibody from the ori-B6.1. Others have reported C1q binding differences in antibodies with similar binding affinities, yet these differ in only the V region (24, 25). Based on our evidence that differences in the VH region are not related to affinity, but may affect complement activation, we hypothesize that the hypermutation potential associated with the B6.1 hybridoma leads to reduced ability of the antibody to fix complement. As others have shown the importance of the variable region of IgM antibodies for enabling the so-called (Fc)5 disc to fix C1q (13), we conclude that the continuity of domain interaction from Fab to Fc is probably an important factor in exposing C1q binding sites and enabling complement activation.
In summary, our work demonstrates that an assessment of protective mannan-specific antibodies against candidiasis must include more than a consideration of specificity and antibody affinity. We also provided insights into a mechanism by which monoclonal antibodies specific for short-chain, beta-linked oligomannosides may lose their protective potential against candidiasis. Finally, when considering antibody constructs for protection against candidiasis, both specificity and complement-fixing activities are of critical concern. Whether human antimannan antibodies play similar roles in patient defense against candidiasis remains to be proven, but it is of interest that, in a recent paper, a human recombinant antibody specific for mannan shows many similarities to protective antibodies that we have described and the recombinant antibody is protective in mice (51).
Acknowledgments
This research was supported by the National Institutes of Health grants RO1 AI24912 and PO1 AI061537.
We thank David Bundle for supplying us with synthetic beta-linked oligomannosides. We are also grateful to technician Melissa Quick, who was involved in initial sequencing experiments, and to Tom Kozel, who provided useful advice on complement fixation assays. We also acknowledge the excellent technical assistance of Sheila Berry and Marcia Riesselman that led to the isolation of hybridoma clone G11.2.
Editor: A. Casadevall
REFERENCES
- 1.Anaissie, E., and G. P. Bodey. 1989. Nosocomial fungal infections. Old problems and new challenges. Infect. Dis. Clin. N. Am. 3:867-882. [PubMed]
- 2.Berenguer, J., M. Buck, F. Witebsky, F. Stock, P. A. Pizzo, and T. J. Walsh. 1993. Lysis-centrifugation blood cultures in the detection of tissue-proven invasive candidiasis. Diagn. Microbiol. Infect. Dis. 17:103-109. [DOI] [PubMed] [Google Scholar]
- 3.Bodey, G. P. 1988. The emergence of fungi as major hospital pathogens. J. Hosp. Infect. 11:411-426. [DOI] [PubMed] [Google Scholar]
- 4.Bromuro, C., A. Torosantucci, P. Chiani, S. Conti, L. Polonelli, and A. Cassone. 2002. Interplay between protective and inhibitory antibodies dictates the outcome of experimentally disseminated candidiasis in recipients of a Candida albicans vaccine. Infect. Immun. 70:5462-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll, W. L., E. Mendel, and S. Levy. 1988. Hybridoma fusion cell lines contain an aberrant kappa transcript. Mol. Immunol. 25:991-995. [DOI] [PubMed] [Google Scholar]
- 6.Casadevall, A. 1995. Antibody immunity and invasive fungal infections. Infect. Immun. 63:4211-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadevall, A., W. Cleare, M. Feldmesser, A. Glatman-Freedman, D. L. Goldman, T. R. Kozel, N. Lendvai, J. Mukherjee, L.-A. Pirofski, J. Rivera, A. L. Rosas, M. D. Scharff, P. Valadon, K. Westin, and Z. Zhong. 1998. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob. Agents Chemother. 42:1437-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casadevall, A., E. Dadachova, and L.-A. Pirofski. 2004. Passive antibody therapy for infectious diseases. Nature Rev. 2:695-703. [DOI] [PubMed] [Google Scholar]
- 9.Casadevall, A., and L.-A. Pirofski. 2004. New concepts in antibody-mediated immunity. Infect. Immun. 72:6191-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clackson, T., H. R. Hoogenboom, A. D. Griffiths, and G. Winter. 1991. Making antibody fragments using phage display libraries. Nature 352:624-628. [DOI] [PubMed] [Google Scholar]
- 11.Cutler, J. E. 2005. Defining criteria for anti-mannan antibodies to protect against candidiasis. Curr. Mol. Med. 5:383-392. [DOI] [PubMed] [Google Scholar]
- 12.de Mattos Grosso, D., S. R. de Almeida, M. Mariano, and J. D. Lopes. 2003. Characterization of gp70 and anti-gp70 monoclonal antibodies in Paracoccidioides brasiliensis pathogenesis. Infect. Immun. 71:6534-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinstein, A., N. Richardson, and M. J. Taussig. 1986. Immunoglobulin flexibility in complement activation. Immunol. Today 7:169-174. [DOI] [PubMed] [Google Scholar]
- 14.Fidel, P. L., M. E. Lynch, and J. D. Sobel. 1993. Candida-specific cell-mediated immunity is demonstrable in mice with experimental vaginal candidiasis. Infect. Immun. 61:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher-Hoch, S. P., and L. Hutwagner. 1995. Opportunistic candidiasis: an epidemic of the 1980s. Clin. Infect. Dis. 21:897-904. [DOI] [PubMed] [Google Scholar]
- 16.Han, Y., and J. E. Cutler. 1995. Antibody response that protects against disseminated candidiasis. Infect. Immun. 63:2714-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han, Y., and J. E. Cutler. 1997. Assessment of a mouse model of neutropenia and the effect of an anti-candidiasis monoclonal antibody in these animals. J. Infect. Dis. 175:1169-1175. [DOI] [PubMed] [Google Scholar]
- 18.Han, Y., T. Kanbe, R. Cherniak, and J. E. Cutler. 1997. Biochemical characterization of Candida albicans epitopes that can elicit protective and nonprotective antibodies. Infect. Immun. 65:4100-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han, Y., T. R. Kozel, M. X. Zhang, R. S. MacGill, M. C. Carroll, and J. E. Cutler. 2001. Complement is essential for protection by an IgM and an IgG3 monoclonal antibody against experimental hematogenously disseminated candidiasis. J. Immunol. 167:1550-1557. [DOI] [PubMed] [Google Scholar]
- 20.Han, Y., R. P. Morrison, and J. E. Cutler. 1998. A vaccine and monoclonal antibodies that enhance mouse resistance to Candida albicans vaginal infection. Infect. Immun. 66:5771-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han, Y., M. H. Riesselman, and J. E. Cutler. 2000. Protection against candidiasis by an immunoglobulin G3 (IgG3) monoclonal antibody specific for the same mannotriose as an IgM protective antibody. Infect. Immun. 68:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han, Y., M. A. Ulrich, and J. E. Cutler. 1999. Candida albicans mannan extract-protein conjugates induce a protective immune response against experimental candidiasis. J. Infect. Dis. 179:1477-1484. [DOI] [PubMed] [Google Scholar]
- 23.Hartman, A. B., and S. Rudikoff. 1984. VH genes encoding the immune response to beta-(1,6)-galactan: somatic mutation in IgM molecules. EMBO J. 3:3023-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horgan, C., K. Brown, and S. H. Pincus. 1990. Alteration in H chain V region affects complement activation by chimeric antibodies. J. Immunol. 145:2527-2532. [PubMed] [Google Scholar]
- 25.Horgan, C., K. Brown, and S. H. Pincus. 1992. Effect of H chain V region on complement activation by immobilized immune complexes. J. Immunol. 149:127-135. [PubMed] [Google Scholar]
- 26.Hostetter, M. K. 1996. New insights into candidal infections. Adv. Pediatr. 43:209-230. [PubMed] [Google Scholar]
- 27.Idusogie, E. E., P. Y. Wong, L. G. Presta, H. Gazzano-Santoro, K. Totpal, M. Ultsch, and M. G. Mulkerrin. 2001. Engineered antibodies with increased activity to recruit complement. J. Immunol. 166:2571-2575. [DOI] [PubMed] [Google Scholar]
- 28.Klaus, G. G. B., M. B. Pepys, K. Kitajima, and B. A. Askonas. 1979. Activation of mouse complement by different classes of mouse antibody. Immunology 38:687-695. [PMC free article] [PubMed] [Google Scholar]
- 29.Li, R.-K., and J. E. Cutler. 1991. A cell surface/plasma membrane antigen of Candida albicans. J. Gen. Microbiol. 137:455-464. [DOI] [PubMed] [Google Scholar]
- 30.Matthews, R. C., G. Rigg, S. Hodgetts, T. Carter, C. Chapman, C. Gregory, C. Illidge, and J. Burnie. 2003. Preclinical assessment of the efficacy of mycograb, a human recombinant antibody against fungal HSP90. Antimicrob. Agents Chemother. 47:2208-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miletic, V. D., and M. M. Frank. 1995. Complement-immunoglobulin interactions. Curr. Biol. 7:41-47. [DOI] [PubMed] [Google Scholar]
- 32.Mukherjee, J., G. Nussbaum, M. D. Scharff, and A. Casadevall. 1995. Protective and nonprotective monoclonal antibodies to Cryptococcus neoformans originating from one B cell. J. Exp. Med. 181:405-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nitz, M., C.-C. Ling, A. Otter, J. E. Cutler, and D. R. Bundle. 2002. The unique solution structure and immunochemistry of the Candida albicans β-1,2-mannopyranan cell wall antigens. J. Biol. Chem. 277:3440-3446. [DOI] [PubMed] [Google Scholar]
- 34.Orlandi, R., D. H. Gussow, P. T. Jones, and G. Winter. 1989. Cloning immunoglobulin variable domains for expression by the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 86:3833-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polonelli, L., W. Magliani, S. Conti, L. Bracci, L. Lozzi, P. Neri, D. Adriani, F. De Bernardis, and A. Cassone. 2003. Therapeutic activity of an engineered synthetic killer antiidiotypic antibody fragment against experimental mucosal and systemic candidiasis. Infect. Immun. 71:6205-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogozin, I. B., and M. Diaz. 2004. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 172:3382-3384. [DOI] [PubMed] [Google Scholar]
- 37.Rogozin, I. B., and N. A. Kolchanov. 1992. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta 1171:11-18. [DOI] [PubMed] [Google Scholar]
- 38.Schaberg, D. R., D. H. Culver, and R. P. Gayner. 1991. Major trends in the microbial etiology of nosocomial infection. Am. J. Med. 16:72S-75S. [DOI] [PubMed] [Google Scholar]
- 39.Schäble, K. F., R. Thiebe, A. Bensch, J. Brensing-Küppers, V. Helm, T. Kirschbaum, R. Lamn, M. Ohnrich, S. Pourrajabi, F. Röschenthaler, J. Schwendinger, D. Wichelhaus, I. Zocher, and H. G. Zachau. 1999. Characteristics of the immunoglobulin Vκ genes, pseudogenes, relics and orphons in the mouse genome. Eur. J. Immunol. 29:2082-2086. [DOI] [PubMed] [Google Scholar]
- 40.Shibata, N., M. Arai, E. Haga, T. Kikuchi, M. Najima, T. Satoh, H. Kobayashi, and S. Suzuki. 1992. Structural identification of an epitope of antigenic factor 5 in mannans of Candida albicans NIH B-792 (serotype B) and J-1012 (serotype A) as beta-1,2-linked oligomannosyl residues. Infect. Immun. 60:4100-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shibata, N., K. Hisamichi, T. Kikuchi, H. Kobayashi, Y. Okawa, and S. Suzuki. 1992. Sequential nuclear magnetic resonance assignment of β-1,2-linked mannooligosaccharides isolated from the phosophomannan of the pathogenic yeast Candida albicans NIH B-792 strain. Biochemistry 31:5680-5686. [DOI] [PubMed] [Google Scholar]
- 42.Spira, G., A. Bargellesi, J.-L. Teillaud, and M. D. Scharff. 1984. The identification of monoclonal class switch variants by sib selection and an ELISA assay. J. Immunol. Methods 74:307-315. [DOI] [PubMed] [Google Scholar]
- 43.Tachibana, H., N. Matsumoto, X.-J. Cheng, H. Tsukamoto, and E. Yoshihara. 2004. Improved affinity of a human anti-Entamoeba histolytica Gal/Ga1NAc lectin Fab fragment by a single amino acid modification of the light chain. Clin. Diagn. Lab. Immunol. 11:1085-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor, B., J. J. Wright, S. Arya, D. E. Isenman, M. J. Shulman, and R. H. Painter. 1994. C1q binding properties of monomer and polymer forms of mouse IgM μ-chain variants. J. Immunol. 153:5303-5313. [PubMed] [Google Scholar]
- 45.Tollemar, J., N. Gross, N. Dolgiras, C. Jarstrand, O. Ringden, and L. Hammarstrom. 1999. Fungal prophylaxis by reduction of fungal colonization by oral administration of bovine anti-Candida antibodies in bone marrow transplant recipients. Bone Marrow Transplant. 23:283-290. [DOI] [PubMed] [Google Scholar]
- 46.Torosantucci, A., C. Bromuro, P. Chiani, F. DeBernardis, F. Berti, C. Galli, F. Norelli, C. Bellucci, L. Polonelli, P. Costantino, R. Rappuoli, and A. Cassone. 2005. A novel glyco-conjugate vaccine against fungal pathogens. J. Exp. Med. 202:597-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voss, A., J. L. M. L. le Noble, F. M. Verduyn Lunel, N. A. Foudraine, and J. F. G. M. Meis. 1997. Candidemia in intensive care unit patients: risk factors for mortality. Infection 25:8-10. [DOI] [PubMed] [Google Scholar]
- 48.Wagner, S. D., C. Milstein, and M. S. Neuberger. 1995. Codon bias targets mutation. Nature 376:732. [DOI] [PubMed] [Google Scholar]
- 49.Wenzel, R. P. 1995. Nosocomial candidemia: risk factors and attributable mortality. Clin. Infect. Dis. 20:1531-1534. [DOI] [PubMed] [Google Scholar]
- 50.Wright, J. F., M. J. Shulman, D. E. Isenman, and R. H. Painter. 1988. C1 binding by murine IgM. J. Biol. Chem. 263:11221-11226. [PubMed] [Google Scholar]
- 51.Zhang, M. X., M. C. Bohlman, C. Itatani, D. R. Burton, P. W. H. I. Parren, S. C. St. Jeor, and T. R. Kozel. 2006. Human recombinant antimannan immunoglobulin G1 antibody confers resistance to hematogenously disseminated candidiasis in mice. Infect. Immun. 74:362-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, M. X., and T. R. Kozel. 1998. Mannan-specific immunoglobulin G antibodies in normal human serum accelerate binding of C3 to Candida albicans via the alternative complement pathway. Infect. Immun. 66:4845-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]