Abstract
We previously proposed a mechanism for control of Rubisco expression and assembly during oxidative stress in Chlamydomonas reinhardtii. The N terminus of the large subunit (LSU) comprises an RNA recognition motif (RRM) that is normally buried in the protein, but becomes exposed under oxidizing conditions when the glutathione pool shifts toward its oxidized form. Thus, de novo translation and assembly of Rubisco LSU stop with similar kinetics and the unpaired small subunit (SSU) is rapidly degraded. Here we show that the structure of the N-terminal domain is highly conserved throughout evolution, despite its relatively low sequence similarity. Furthermore, Rubisco from a broad evolutionary range of photosynthetic organisms binds RNA under oxidizing conditions, with dissociation constant values in the nanomolar range. In line with these observations, oxidative stress indeed causes a translational arrest in land plants as well as in Rhodospirillum rubrum, a purple bacterium that lacks the SSU. We highlight an evolutionary conserved element located within α-helix B, which is located in the center of the RRM and is also involved in the intramolecular interactions between two LSU chains. Thus, assembly masks the N terminus of the LSU hiding the RRM. When assembly is interrupted due to structural changes that occur under oxidizing conditions or in the absence of a dedicated chaperone, the N-terminal domain can become exposed, leading to the translational arrest of Rubisco LSU. Taken together, these results support a model by which LSU translation is governed by its dimerization. In the case that regulation of type I and type II Rubisco is conserved, the SSU does not appear to be directly involved in LSU translation.
Rubisco is the enzyme responsible for CO2 fixation during photosynthesis. In land plants and green algae, Rubisco exists as a holoenzyme composed of eight large subunits (LSUs; 55 kD) encoded by the chloroplast large subunit of Rubisco (rbcL) gene, and eight small subunits (SSU; 15–18 kD) produced by a nuclear family of small subunit of Rubisco (rbcS) genes (Spreitzer, 1993). This L8S8 form of the complex is denoted form I (Spreitzer, 1999). SSU precursors are processed during entry into the chloroplast and are then assembled with the LSUs to yield the holoenzyme (approximately 600 kD). The Rubisco protein complex of red algae, which also exists as an L8S8 form, differs from that in land plants and green algae in that both subunits are encoded by the chloroplast genome, and the complex is therefore denoted red Rubisco, as opposed to the green version of land plants and green alga (Spreitzer, 1999). Some prokaryotes and dinoflagellates lack the SSU, and the Rubisco holoenzyme of these organisms, denoted form II, is assembled from only two LSU chains (Tabita, 1999). Several hypotheses have been put forward to explain the role of the SSU but to date none is universally accepted (Spreitzer, 1999).
Expression of the two subunits is highly coordinated, and down-regulation of the SSU by antisense RNA causes a parallel reduction in expression of the LSU (Rodermel et al., 1996). The coordinated expression of subunits that comprise large photosynthetic complexes takes place in the chloroplast, in a process that is designated controlled by epistasy of synthesis (CES; Choquet et al., 2003; Wostrikoff et al., 2004). Thus, in the absence of one subunit in the complex, synthesis of another subunit of the complex decreases (Choquet et al., 2003; Wostrikoff et al., 2004; Minai et al., 2006).
We recently proposed an autoregulatory mechanism for control of Rubisco expression and assembly in Chlamydomonas reinhardtii during light-induced oxidative stress (Cohen et al., 2005). Under oxidizing conditions the glutathione pool shifts toward the oxidized form (GSSG), affecting the redox state of thiol groups on both Rubisco subunits. As a result of these changes, translation of the LSU stops, assembly of new particles ceases, and the SSU is rapidly degraded. While searching for regulatory components that interact with the rbcL transcript we found that under oxidizing conditions, the Rubisco LSU can bind RNA in vitro in a sequence-independent manner and that the N terminus (150 amino acids) is responsible for this activity. Under normal conditions, the N terminus is buried in the folded protein, but upon oxidation of the protein it becomes exposed and will bind any RNA in the vicinity (Yosef et al., 2004). Indeed, under oxidizing conditions, the rbcL transcript remains in association with ribosomes but shifts toward the monosomal fraction.
The role of the SSU in this proposed mechanism has not been fully elucidated but it can follow several different routes. Oxidative stress can trigger SSU degradation, thus leading to translational arrest of the LSU. Alternatively, oxidative stress could initially arrest LSU translation, resulting in a rapid degradation of the unassembled SSU. Finally, it may be that both subunits and possibly their chaperones are subject to conformational changes that prevent their assembly; hence, translation of the LSU stops and the SSU is degraded. Under oxidizing conditions, translational arrest of the LSU and inhibition of the assembly process occur with similar kinetics, suggesting that the two processes are indeed tightly coupled (Cohen et al., 2005). With the aims of obtaining a better understanding of the regulatory mechanism and of following its evolutionary origin, we examined the pattern of LSU translation under oxidizing conditions in land plants, as well as in the photosynthetic purple bacterium Rhodospirillum rubrum, which expresses form II Rubisco. In all three organisms the LSU shows CES behavior, as its translation stops under oxidizing conditions. This suggests that a phylogenetically conserved regulatory mechanism is involved in regulation of Rubisco in all photosynthetic organisms.
RESULTS
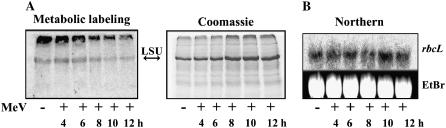
Oxidative Stress Causes a Translational Arrest of Rubisco LSU in Tobacco and Spinach without Affecting the Level of the rbcL Transcript
We sought to generalize our previous observations in C. reinhardtii that light-induced oxidative stress leads to a dramatic, though transient, arrest in Rubisco LSU translation that is tightly coordinated with cessation of its assembly (Cohen et al., 2005). As a first step, we examined the translation pattern of Rubisco LSU in tobacco (Nicotiana tabacum) leaves in response to oxidative stress induced by methyl viologen (MeV). In these experiments, nascently synthesized proteins were labeled with a pulse of radioactive amino acids. Incorporation of the radiolabel was identical under all conditions, confirming that the cells were viable throughout the treatment with MeV that extended up to 12 h. Translation shut down of Rubisco LSU occurred already after 8 h of incubation with MeV, as compared with the untreated control and with leaf discs exposed to MeV for a shorter time, 4 and 6 h (Fig. 1A). At 16 h there was a decrease in the overall nascent incorporation of radiolabeled material into proteins, accompanied with severe bleaching of the cells (data not shown). Indeed, a complete chlorotic damage was shown for leaf discs of tobacco after 18 h incubation with 2 μm MeV (Youssefian et al., 2001). The translational arrest could not have originated from a potential reduction in the RNA level, since the rbcL transcript remained unchanged upon exposure to MeV for similar time periods (Fig. 1B). Thus, oxidative stress of tobacco leaves resulted in a reduction in the de novo synthesis of Rubisco LSU, as was previously shown for C. reinhardtii (Shapira et al., 1997). A similar labeling pattern was obtained for spinach (Spinacia oleracea) leaf discs (data not shown).
Figure 1.
Oxidative stress causes a translational arrest of Rubisco LSU in land plants without affecting mRNA levels. A, The effect of oxidative stress on protein synthesis was monitored by metabolic labeling of tobacco leaf discs under normal conditions and following exposure to (2 μm) MeV for time periods up to 12 h. Equal protein loads were analyzed on 12% SDS gels, which were then subjected to autoradiography (left section) or Coomassie staining (right section). Migration of Rubisco LSU is indicated with an arrow. B, Northern analysis of the rbcL transcript in leaves (of plants grown under normal conditions) that were subjected to oxidative stress as described in A.
The Structure of the N-Terminal Domain of Rubisco LSU Is Conserved throughout Evolution Despite Its Relatively Variable Sequence
We recently proposed that exposure of the N-terminal domain (1–150 amino acids) of Rubisco LSU from C. reinhardtii to oxidizing conditions, and the consequent binding of RNA in a sequence-independent manner (Yosef et al., 2004), plays a role in the regulation of Rubisco expression. The N terminus of Rubisco LSU is not involved in CO2 fixation, as the active site of the enzyme lies in the C-terminal domain, which has the structure of an eight-stranded α/β barrel (TIM barrel). However, the N terminus of one subunit associates with the C terminus of the neighboring chain and is therefore involved in the assembly process (Schneider et al., 1986, 1990a).
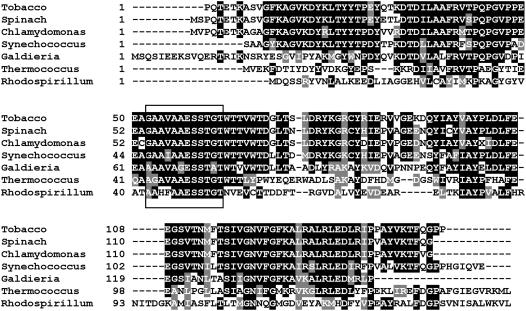
To examine the phylogenetic conservation of RNA-binding activity, we aligned the sequences of the N-terminal domain (positions 1–150) of Rubisco LSU from tobacco, spinach, Synechococcus PCC 6301, Galdiera partita, Thermococcus kodakaraensis, and Rhodospirillum rubrum (Table I). The highest degree of identity (80%–87%) was observed between the green, form I Rubisco polypeptides from tobacco, spinach, Synechococcus PCC 6301, and C. reinhardtii. Red, form I Rubisco LSU from G. partita showed a lower similarity to the polypeptides mentioned above (56.7%) and the lowest conservation values were measured for the archeal protein from T. kodakaraensis (38.2%) and for the bacterial protein from R. rubrum (29.6%). Despite the low conservation between the archeal protein and the green Rubisco, the alignment revealed a distinctive and highly conserved sequence element that corresponded to positions 54 to 65 in form I Rubisco LSUs and to positions 43 to 54 and 42 to 53 in the LSU chains of T. kodakaraensis and R. rubrum, respectively (Fig. 2). An examination of the three-dimensional structure of Rubisco (Schneider et al., 1986, 1990a) shows that some of the residues found in this hydrophobic conserved sequence element, for example Glu-48, Ser-50, and Thr-51 in the N terminus of one subunit, form hydrophobic contacts with the C terminus of the second subunit.
Table I.
Sequence conservation of the LSU N terminus among different photosynthetic organisms
| Species | Homology with the N-Terminal Domain (1–150) of C. reinhardtii LSU | Homology with the C. reinhardtii Rubisco LSU (1–475) |
|---|---|---|
| Tobacco | 88.5% | 84.4% |
| Spinach | 86.7% | 87.7% |
| Synechococcus PCC6301 | 74.6% | 81.6% |
| G. partita | 59.2% | 56.7% |
| T. kodakaraensis | 41.5% | 38.2% |
| R. rubrum | 38.6% | 29.6% |
Figure 2.
Sequence conservation of the N-terminal domain of Rubisco. Sequence of the N-terminal domain (amino acids 1–150) of Rubisco of C. reinhardtii, tobacco, Spinach, Synechococcus PCC6301, G. partita, T. kodakaraensis, and R. rubrum were aligned using ClustalW. Identical and similar amino acids are shaded with black and gray backgrounds, respectively. The highly conserved sequence element is marked with a black rectangle.
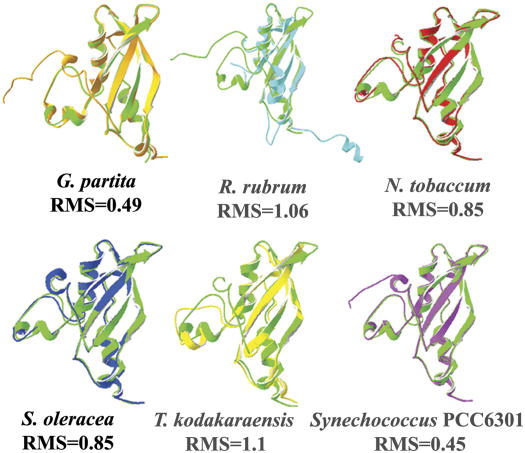
To evaluate whether the sequence variations in the N terminus could cause alterations in the overall structure, we superimposed the published structures of the N-terminal domains from each organism on that of C. reinhardtii (Taylor et al., 2001). Figure 3 shows the individually superimposed structures, based on alignment of the Cα atoms in all N-terminal domains, and their positional root mean square (RMS) deviations. The highest structural resemblance to the LSU of C. reinhardtii was observed for the LSU chains of the protein from the cyanobacterium Synechococcus PCC 6301 (Newman and Gutteridge, 1993; RMS of 0.45) and the protein from the rhodophyte G. partita (Sugawara et al., 1999; RMS of 0.49). A high structural similarity (RMS of 0.85) was observed for the spinach (Taylor and Andersson, 1996) and tobacco (Duff et al., 2000) proteins. The RMS values of proteins from organisms as remote as C. reinhardtii, R. rubrum (Schneider et al., 1990b), and T. kodakaraensis (Kitano et al., 2001) also indicated a strong structural resemblance that did not exceed 1.06 and 1.1 Å for LSU, respectively. These findings are in agreement with previous reports that emphasize the structural resemblance between Rubisco proteins from R. rubrum (Schneider et al., 1990a) and spinach (Taylor and Andersson, 1996), showing a similar range of RMS values (Schneider et al., 1990a). Thus, despite the significant differences in the amino acid sequences of the Rubisco N-terminal domains, the structural alignment revealed a remarkable conservation throughout evolution. The typical basic pattern of the βαββαβ structure was found in all the N termini of the structures, and as previously reported (Yosef et al., 2004) it is typical of the RNA recognition motif (Nagai, 1996; Siomi and Dreyfuss, 1997). The high degree of evolutionary conservation observed in the structure of the N terminus suggests that this region could play a similar role in different photosynthetic organisms, possibly in the complex regulation of the assembly process and subunit expression.
Figure 3.
The structure of the N-terminal domain of Rubisco is highly conserved among different photosynthetic organisms. A computerized alignment of the three-dimensional structures of Rubisco from different organisms was performed using the published structures for spinach (1AUS, blue), tobacco (1EJ7, red), G. partita (a rhodophyte, 1BWV, orange), T. kodakaraensis (a photosynthetic archeaon, 1GEH, yellow), R. rubrum (a purple bacterium, 5RUB, cyan), and from Synechococcus PCC6301 (a cyanobacterium, 1RBL, purple). All structures were compared using SPDB Viewer, with the structure of C. reinhardtii (a green alga, 1GK8, green). The RMS deviation values of each alignment appear below each superimposed structure.
RNA-Binding Activity of Purified Rubisco LSU from Different Organisms
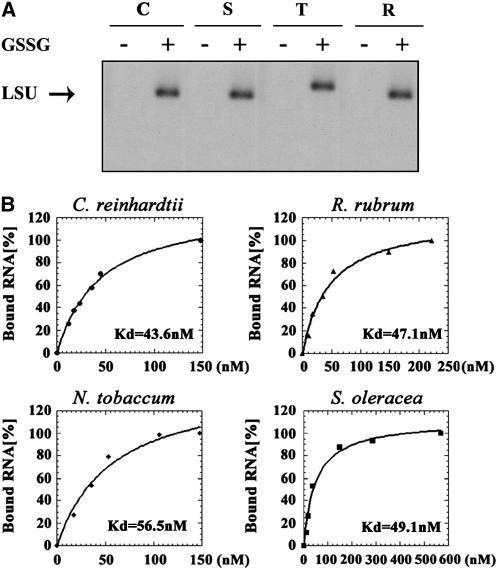
In view of the highly conserved structure of the N terminus of Rubisco LSU, we characterized the kinetics of RNA-binding activity of Rubisco from species of different phyla, i.e. from tobacco and from spinach (Magnoliophyta), from C. reinhardtii (Chlorophyta), and from R. rubrum (Proteobacteria), the latter being a phylogenetically remote photosynthetic prokaryote, with the lowest degree of sequence conservation for Rubisco LSU. In these experiments, purified Rubisco was incubated with radiolabeled RNA under oxidizing and normal conditions, i.e. with or without GSSG, respectively. In keeping with our previous observations for C. reinhardtii, RNA-binding activity was observed only under oxidizing conditions for purified Rubisco from land plants and from the species purple bacterium (Fig. 4A).
Figure 4.
RNA-binding activity under oxidizing conditions of purified Rubisco is conserved throughout evolution. A, Purified Rubisco proteins from C. reinhardtii (C), spinach (S), tobacco (T), and R. rubrum (R) were used in UV cross-linking experiments performed in the presence or absence of GSSG. B, To determine the Kd of RNA binding, increasing concentrations of purified Rubisco proteins (10–500 nm) were UV cross-linked under oxidizing conditions to a random transcript (250 nts) that was transcribed from the multicloning site of pBluescript. Following RNase A treatment, the radiolabeled proteins were separated over 15% SDS-PAGE and their labeling was monitored using a phosphorimager. The relative amount of the bound radioactivity was plotted against the protein concentration using Kaleidograph software, and the Kd was calculated for each polypeptide.
As a measure of the binding affinities of different Rubisco holoenzymes to RNA, we determined the dissociation constant (Kd) of purified Rubisco from tobacco, spinach, C. reinhardtii, and R. rubrum. The binding assays were performed in the presence of radiolabeled RNA that was synthesized in vitro from the multiple cloning site of pBluescript (approximately 250 nts; Fig. 4, A and B), and also the 5′ untranslated region (UTR) of rbcL RNA of C. reinhardtii (100 nts; data not shown). The Kd values of all Rubisco holoenzymes tested were similar for the two substrates, despite the sequence variations in the N terminus, but in line with the high degree of structural conservation among them. Results with the RNA synthesized on the pBluescript template ranged between 43.6 and 56.5 nm for all the proteins (Fig. 4B). These values are consistent with our previous report on RNA binding by Rubisco LSU and by the recombinant N terminus of C. reinhardtii (Yosef et al., 2004).
Exposure of R. rubrum to Atmospheric Air Causes the Translational Arrest of Rubisco LSU
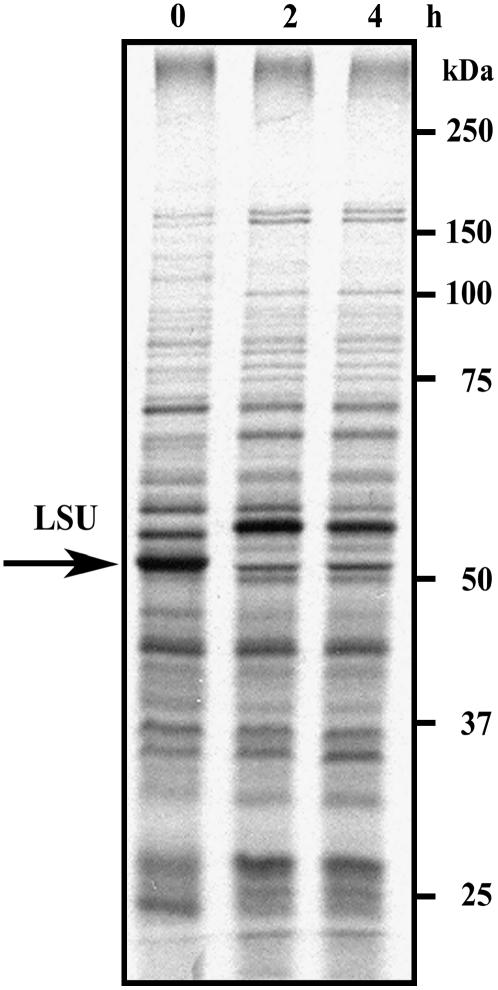
Rubisco of R. rubrum consists of two LSU chains (L2) and does not contain an SSU. We were thus interested to examine whether in the absence of the SSU, LSU synthesis is subject to regulation by oxidative stress, as observed for C. reinhardtii and land plants. R. rubrum is photosynthetic only when grown under anaerobic conditions (Anderson and Fuller, 1967). Exposure of an anaerobic culture of R. rubrum to atmospheric air reduced the activity of Rubisco to less than 20% within 12 h, while the steady-state level of the enzyme remained constant (Cook and Tabita, 1988). Extensive degradation was observed only after extended exposure to aerobic conditions (48 to 72 h). However, when cultures that had been preexposed to atmospheric air were shifted back to anaerobic conditions under an argon atmosphere, Rubisco activity increased rapidly. The authors suggested that this increase was due to de novo synthesis of the enzyme, although other possibilities cannot be excluded. To examine the influence of aerobic conditions on LSU translation in R. rubrum, cells were shifted from anaerobic to aerobic conditions for time periods of 0, 2, and 4 h, and then pulse labeled for 5 min. Total cell proteins were extracted and separated over SDS-PAGE. Migration of the LSU polypeptide was identified by using specific antibodies, and the autoradiograms were aligned with the colored blots. Purified Rubisco was also separated on the same blots that were reacted with the specific antibodies and used as a marker for the exact identification of the protein (data not shown). As shown in Figure 5, exposure to an oxidizing atmosphere terminated Rubisco translation within 2 h, the dramatic effect on translation being similar to that observed in photo-inhibited C. reinhardtii cells (Shapira et al., 1997).
Figure 5.
Nascent synthesis of Rubisco LSU in R. rubrum is inhibited by exposure to atmospheric air. Pulse labeling of the photosynthetic bacterium R. rubrum was performed on midlog phase cultures growing at 30°C under anaerobic conditions and after exposure to atmospheric air. Pulses of 5 min were given at intervals of 0, 2, and 4 h with 75 μCi of [35S]Met cell-labeling mix. Equal protein loads that also contained equal amounts of incorporated radioactivity were analyzed over 7.5% to 12.5% SDS-PAGE. The gels were blotted and the blots were reacted with antibodies against global Rubisco. The colored developed blots were aligned with the autoradiogram that is shown in the figure. Rubisco LSU is marked with an arrow.
DISCUSSION
In this study, we showed that the regulatory mechanism that directs translational regulation under extreme conditions is conserved throughout evolution. We found that despite extensive variations in the sequence of the N terminus among different photosynthetic organisms, significant conservation of its structure was revealed. Structural conservation is to be expected at the active site of Rubisco, which is located at the C-terminal TIM barrel domain of the LSU. A similar finding for the N terminus could suggest that it too plays an important and conserved role, possibly in the regulation of Rubisco expression.
We show here that the RNA-binding activity of Rubisco LSU is conserved throughout evolution. UV cross-linking experiments performed with purified Rubisco from land plants (tobacco and spinach) and from R. rubrum showed similar patterns, namely, that RNA binding occurs only under oxidizing conditions, with Kd values in the same nanomolar range as the value calculated for Rubisco from C. reinhardtii. These findings are in line with the high structural conservation observed for Rubisco LSU, despite the sequence variability in the N-terminal domain.
It has previously been shown that the SSU is not required for assembly of Aspergillus nidulans LSU chains into the L8 octomer core in Escherichia coli and that the SSU could be added to preformed cores to generate an active enzyme. It was therefore suggested that a face-to-bottom structure of two LSU chains could be the basic and common step involved in Rubisco assembly (Goloubinoff et al., 1989). Similar results were also reported for a cyanobacterial LSU that was expressed and assembled in E. coli in the absence of the SSU, generating oligomeric particles (van der Vies et al., 1986). It appears that in these cases the SSU was not required for assembly or for solubility of Rubisco, although the intermolecular binding of the L8 structure was weaker than that of the L8S8 form (Andrews, 1988). Indeed, the SSU is a late evolutionary addition, as the primitive L2 form of Rubisco found in some prokaryotes and dinoflagellates does not contain an SSU (Spreitzer, 2003). Here we show that similar to C. reinhardtii, exposure of R. rubrum to an oxidizing environment results in a translational arrest of Rubisco LSU. Since R. rubrum encodes for form II Rubisco, which does not contain an SSU chain, it seems that preventing dimerization of the LSU is sufficient to block its translation. Indeed, according to the model that we propose (shown in Fig. 6), when LSU chains cannot pair, their N terminus is unveiled, triggering RNA binding and preventing the ribosome from advancing along the rbcL transcript. However, when two LSU molecules form a dimer, the N terminus of one chain is masked due to its association with the C terminus of the second chain, thus disabling the RNA-binding activity. We propose that the N terminus must be masked at all times, either by interaction of the nascent polypeptide with chaperones, or by dimerization of two LSU molecules as part of the assembly process. Oxidative stress causes structural alterations that most probably prevent assembly (Cohen et al., 2005), leading to exposure of the N terminus and blocking translation.
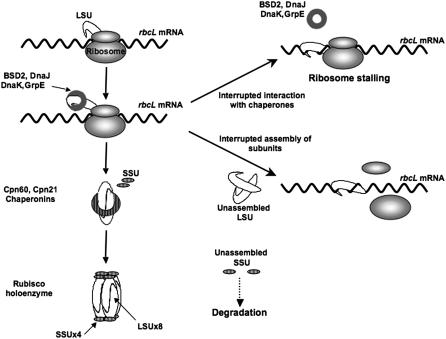
Figure 6.
A model for the proposed regulation of Rubisco. Rubisco LSU chains are synthesized in the chloroplast and interact with the dnaK/dnaJ/grpE chaperone complex that maintains the chains in their unfolded state. During assembly LSU subunits are attached in a head to tail fashion. Assembly of the mature L8S8 holoenzyme is promoted by the chaperonin system. Conformational changes induced by oxidative stress either in Rubisco subunits or in the related chaperones inhibit assembly by preventing interactions between the LSU subunits or between the LSU and the chaperone system. Under these conditions, the N-terminal domain of the LSU is accessible to any RNA in its vicinity, causing ribosome stalling. Unpaired SSU chains are rapidly degraded.
Sequence comparison of the N-terminal domain of LSUs from different organisms reveals a highly conserved region of 12 amino acids (corresponding to positions 54–65 in the C. reinhardtii protein; see Fig. 1) found in all photosynthetic organisms, from purple bacteria to land plants. Conserved amino acids that are located in this hydrophobic region have been implicated in the interactions between the N terminus of one subunit and the C terminus of a counterpart chain, allowing the formation of the L2 dimer in R. rubrum (Schneider et al., 1990a). Amino acids such as Glu-48, Ser-50, and Thr-51 of R. rubrum LSU are found within a distance of 3 Å from amino acids located in the C terminus of the second subunit (Pro-176, Lys-168, and Leu-169) and are therefore likely to create the intermolecular interactions between the two subunits. Furthermore, in C. reinhardtii, a natural Gly to Asp substitution at position 54 in the LSU (C. reinhardtii mutant 31-4E) completely eliminated the accumulation of the Rubisco holoenzyme. Based on the crystal structure of spinach LSU, it has been suggested that Gly-54 is likely to be in Van der Waals contact with Cys-84 and Cys-99, which may explain the redox sensitivity of the RNA-binding activity by the N-terminal domain (Thow and Spreitzer, 1992). Moreover, pseudoreversion of the Chlamydomonas mutant 31-4E to a photosynthetic strain occurred by replacement of the Asp residue with Ala or Val (Spreitzer et al., 1995). Interestingly, Ala occupies this position in several photosynthetic organisms, such as G. partita, T. kodakaraensis, and R. rubrum. Gly-54 is located within α-helix B, which is part of the βαββαβ ferredoxin-like domain at the N terminus that is predicted to be responsible for the RNA-binding activity (Yosef et al., 2004). During translation, this domain is most probably masked by chaperones that interact with the nascent chains to maintain them in an unfolded state until their assembly. Once folded and assembled even as the initial dimer, the N terminus is buried within the protein. A putative chaperone that could be involved in the process is the BUNDLE SHEATH DEFECTIVE2 (BSD2) homolog, a DnaJ-like protein that has been shown to be necessary for Rubisco expression in maize (Zea mays; Brutnell et al., 1999). Exposure to oxidizing conditions that affect the redox state of thiol groups on Rubisco subunits as well as on their chaperones can prevent the chaperone activity and hence the assembly process due to conformational changes (Cohen et al., 2005). Indeed, BSD2 contains four CXXC elements at conserved positions, indicating that it could be redox sensitive.
Expression of both the LSU and SSU depends on their mutual compatibility to assemble. The SSU stabilizes the holoenzyme complex, generating fully active and functional Rubisco particles. However, prokaryotes and eukaryotes have different requirements for an SSU to form an active complex. While the cyanobacterial LSU can assemble into a fully active particle even in the absence of an endogenous SSU (Andrews, 1988), silencing of the SSU in tobacco (by antisense RNA) prevented Rubisco assembly and synthesis of the LSU was reduced (Rodermel et al., 1996). The difference between prokaryotes and eukaryotes is further reflected by the inability of tobacco LSU to assemble into a soluble octomeric structure in E. coli, unlike its cyanobacterial counterpart. Thus, it is difficult to individually express the LSU from land plants or green algae in E. coli. The basis for this difference is not yet clear, but we assume that in eukaryotes the initial assembly products, namely L2 or L8, are unstable without the SSU and translation of LSU chains alone is therefore restricted. Since assembly seems to control LSU expression in both forms I and II of Rubisco, it does not seem likely that the SSU has a direct effect on LSU translation. Indeed, the L2 form of Rubisco from R. rubrum can replace the endogenous L8S8 holoenzyme and in tobacco plants it can fully support photosynthetic growth (Whitney and Andrews, 2001a).
In higher plants and in green algae, expression of subunits that are part of large protein complexes is highly coordinated by CES (Choquet et al., 1998, 2001). The basis for this control system is that elimination of one subunit reduces the expression of other subunits in the complex by their translational arrest, or by degradation of unassembled subunits. It was shown, for example, for the cytochrome b6f complex, that translation of cytochrome f, encoded by the petA gene, is autoregulated by its own C terminus, via an element in the 5′ UTR of the petA mRNA (Choquet et al., 2003). A model was proposed in which this C-terminal domain is shielded in the assembled complex. Once assembly is inhibited, this domain is exposed and translation is inhibited. The de novo synthesis of distinct components of PSII, the reaction center subunits D1 and D2 and the core antenna subunit apoCP47 (encoded by psbA, psbD, and psbB, respectively), also appears to be regulated by CES. Reporter genes cloned under control of the 5′ UTRs of the psbA and psbB genes were used to reveal a cascade of events in which D2 is required for expression of the D1 polypeptide, and the latter is necessary for enabling efficient expression of CP47 (Minai et al., 2006). It now appears that Rubisco also behaves like a CES protein and the regulated expression of its LSU is subject to the negative autoregulation that occurs when assembly is prevented. This pattern of regulation pertains to both forms I and II of Rubisco and may even be observed in the absence of the SSU, as was shown here for R. rubrum. The inhibitory N-terminal RNA-binding domain is masked when the LSU homodimer is created, or alternatively, when a chaperone is bound to the newly synthesized chain prior to its dimerization. However, unlike the CES of proteins that are part of the PSI or PSII complexes, for which no RNA-binding activity was shown, Rubisco LSU can interact directly with RNA. The specificity of this interaction, which exclusively targets rbcL transcripts, could be directed by the spatial distribution of the nascently synthesized LSU chains especially since the rbcL RNA remains associated with ribosomes. In summary, this study supports a model by which LSU translation is governed by its dimerization. In the case that regulation of type I and type II Rubisco is conserved, the SSU does not appear to be directly involved in LSU translation.
MATERIALS AND METHODS
Strains and Growth Conditions for Microorganisms and Plants
Cultures of Chlamydomonas reinhardtii wild-type strain CC-125 were cultivated in 300 mL of high-salt reduced sulfate medium (Schmidt et al., 1985) with constant bubbling of 5% CO2, rotary shaking at 25°C, and illumination with cool-white fluorescent light (70 μmol m−2 s−1).
Rhodospirillum rubrum was cultured in 25-mL screw-cap tubes or filled to the top (anaerobic conditions) at 30°C in Ormerod synthetic medium containing malate and 0.2% ammonium sulfate (Bose, 1963; Tabita and McFadden, 1974). Cultures were illuminated with 60-W soft light bank bulbs. In addition, R. rubrum cultures were inoculated with 20 mL of late logarithmic-phase cells grown in the presence of 0.4% butyrate, a procedure that greatly enhances the synthesis and accumulation of Rubisco (Sarles and Tabita, 1983). The cultures were grown anaerobically at 30°C in 1-L Erlenmeyer flasks.
Wild-type tobacco (Nicotiana tabacum) and spinach (Spinacia oleracea) were grown in a commercial soil mixture for 10 to 12 weeks from germination and fertigated with a commercial nutrient solution (Ecogan). The plants were maintained in a controlled environment chamber at 25°C under a photoperiodic illumination regime of 8 h light and 16 h dark.
Pulse Labeling of Total Cell Proteins in Vivo
In vivo pulse labeling of total cell proteins in tobacco plants was performed essentially as described (Whitney and Andrews, 2001b), with the following modifications: Six leaf discs (1 cm2) taken from young fully developed leaves were immediately immersed in 5 mL of Murashige and Skoog nutrient solution (pH 7.2; Murashige and Skoog, 1962) containing 1 mm NaHCO3 and 0.25% (v/v) Silwet-L77 (Lehle Seeds). To induce oxidative stress, MeV (2 μm) was added to the solution and the leaf discs were vacuum infiltrated for 90 s, followed by illumination at 300 μmol m−2s−1 for up to 12 h. Proteins were radiolabeled (for 20 min) with 400 μCi of 35S cell-labeling mix (AGQ0080, Amersham), followed by vacuum infiltration for 90 s. The leaf discs were then removed, frozen in liquid N2, and ground in 400 μL of 0.1 m Tris-HCl pH 8.0, 0.3 m NaCl, and 5 mm MgCl2. Cell debris was spun down (2 min, 8,000g). Incorporation of the radiolabeled amino acids was determined by trichloroacetic acid precipitation and equal protein loads were separated by 12% SDS-PAGE. The gels were stained, dried, and analyzed by a phosphorimager.
To enhance the expression of Rubisco in R. rubrum the cells were grown anaerobically in the presence of butyrate. A culture of R. rubrum was grown under anaerobic conditions for 48 h and used to inoculate a butyrate-based medium that was further grown for 3 d. Exposure to atmospheric air was achieved by transferring samples (5 mL) of the logarithmic cultures into open 50-mL test tubes, with extensive stirring. These aliquots were labeled by the addition of 75 μCi of 35S cell-labeling mix for 5 min. Labeling was performed both under anaerobic conditions and after exposure of the cultures to atmospheric air for 0, 2, and 4 h. The labeled cells were collected by centrifugation (5 min, 8,000g at 4°C), resuspended in SDS sample buffer, and boiled for 10 min. Equal protein loads were separated by 7.5% to 12.5% SDS-PAGE. The gels were stained, dried, and analyzed by phosphorimager. Migration of Rubisco LSU was monitored on parallel western blots.
Antisera
Affinity-purified rabbit antibodies raised against a synthetic peptide of Rubisco (Agrisera) were used to detect Rubisco forms I and II. Alternatively, Rubisco form I was identified by rabbit antibodies that were raised against the purified holoenzyme from tobacco.
Rubisco Purification
Rubisco from C. reinhardtii (CC-125) was purified as previously described (Cohen et al., 2005). For purification of Rubisco from tobacco and spinach, 15 g of leaves were frozen in liquid N2 immediately upon picking and ground with 50 mm Tris-HCl pH 7.5, 10 mm MgCl2, 10 mm NaHCO3, and 10 mm dithiothreitol in a Polytron-PT-mr2100 for 5 min. The mixture was centrifuged at 4°C for 10 min at 20,000g. A pellet from a 25% to 50% ammonium sulfate cut of the supernatant was dissolved and dialyzed in dialysis buffer containing 50 mm Tris-HCl pH 7.5, 10 mm Mg2Cl, 10 mm NaHCO3, 10 mm dithiothreitol, and 1 mm EDTA. A 3-mL aliquot of the dialyzed proteins was loaded on a 10% to 30% linear gradient of Suc in dialysis buffer. The gradient was centrifuged for 16 h at 164,000g (SW40 rotor, 40,000 rpm in a Beckman LE-80 K ultracentrifuge) at 4°C, and 1-mL fractions were collected. Samples of each fraction were examined by western-blot analysis using anti-Rubisco antibodies. Rubisco form II from R. rubrum was purified as previously described (Luo et al., 2001) and analyzed by western blotting using the same antibodies.
RNA Extraction, Hybridization, and UV Cross-Linking
Oxidative stress was induced by vacuum infiltration of MeV (2 μm) for 90 s, followed by incubation for up to12 h with illumination conditions of 300 μmol m−2s−1. The harvested leaves were immersed immediately in 5 mL of Murashige and Skoog nutrient solution (pH 7.2) containing 10 mm NaHCO3 and 0.25% (v/v) Silwet-L77 (see above). Total RNA was extracted from whole tobacco leaves (0.5–0.7 g) with the EZ-RNA isolation kit (Biological Industries). Northern blots were hybridized with an rbcL probe that was amplified from tobacco genomic DNA using the sense TobaccoLSUfwd (1–21) primer ATGTCACCACAAACAGAGACTAA and the antisense TobaccoLSUrev (1,411–1,434) primer TTACTTATCCAAAACGTCCACTG. UV cross-linking assays were performed as previously described (Yosef et al., 2004).
Acknowledgments
We are grateful to Zippora Gromet-Elhanan, Weizmann Institute of Science, Israel, for providing us with Rhodospirillum rubrum and with instructions on how to grow it. We thank Robert Tabita, Ohio State University, for the antibodies directed against Rubisco form II, and Yedidia Gafni, Agricultural Research Institute, Israel, for providing us with spinach seeds.
This work was supported by the Israel Science Foundation (grant no. 587/02).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Michal Shapira (shapiram@bgu.ac.il).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.079046.
References
- Anderson L, Fuller RC (1967) Photosynthesis in Rhodospirillum rubrum. I. Autotrophic carbon dioxide fixation. Plant Physiol 42: 487–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews TJ (1988) Catalysis by cyanobacterial ribulose-bisphosphate carboxylase large subunits in the complete absence of small subunits. J Biol Chem 263: 12213–12219 [PubMed] [Google Scholar]
- Bose SK (1963) Media for anaerobic growth of photosynthetic bacteria. In H H Gest, A San Petro, LP Vernon, eds, Bacterial Photosynthesis. Antioch Press, Yellow Springs, OH
- Brutnell TP, Sawers RJ, Mant A, Langdale JA (1999) BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell 11: 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Stern DB, Wostrikoff K, Kuras R, Girard-Bascou J, Wollman FA (1998) Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc Natl Acad Sci USA 95: 4380–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Wostrikoff K, Rimbault B, Zito F, Girard-Bascou J, Drapier D, Wollman FA (2001) Assembly-controlled regulation of chloroplast gene translation. Biochem Soc Trans 29: 421–426 [DOI] [PubMed] [Google Scholar]
- Choquet Y, Zito F, Wostrikoff K, Wollman FA (2003) Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell 15: 1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen I, Knopf JA, Irihimovitch V, Shapira M (2005) A proposed mechanism for the inhibitory effects of oxidative stress on Rubisco assembly and its subunit expression. Plant Physiol 137: 738–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook LS, Tabita FR (1988) Oxygen regulation of ribulose 1,5-bisphosphate carboxylase activity in Rhodospirillum rubrum. J Bacteriol 170: 5468–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff AP, Andrews TJ, Curmi PM (2000) The transition between the open and closed states of rubisco is triggered by the inter-phosphate distance of the bound bisphosphate. J Mol Biol 298: 903–916 [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, Gatenby AA, Lorimer GH (1989) GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose biphosphate carboxylase oligomers in Escherichia coli. Nature 337: 44–47 [DOI] [PubMed] [Google Scholar]
- Kitano K, Maeda N, Fukui T, Atomi H, Imanaka T, Miki K (2001) Crystal structure of a novel-type archaeal rubisco with pentagonal symmetry. Structure 9: 473–481 [DOI] [PubMed] [Google Scholar]
- Luo S, Wang ZY, Kobayashi M, Nozawa T (2001) The dimerization of folded monomers of ribulose 1,5-bisphosphate carboxylase/oxygenase. J Biol Chem 276: 7023–7026 [DOI] [PubMed] [Google Scholar]
- Minai L, Wostrikoff K, Wollman FA, Choquet Y (2006) Chloroplast biogenesis of Photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 18: 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Nagai K (1996) RNA-protein complexes. Curr Opin Struct Biol 6: 53–61 [DOI] [PubMed] [Google Scholar]
- Newman J, Gutteridge S (1993) The x-ray structure of Synechococcus ribulose-bisphosphate carboxylase/oxygenase-activated quaternary complex at 2.2-A resolution. J Biol Chem 268: 25876–25886 [PubMed] [Google Scholar]
- Rodermel S, Haley J, Jiang CZ, Tsai CH, Bogorad L (1996) A mechanism for intergenomic integration: abundance of ribulose bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc Natl Acad Sci USA 93: 3881–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarles L, Tabita R (1983) Derepression of the synthesis of D-ribulose 1,5-bisphosphate carboxylase/oxygenase from Rhodospirillum rubrum. J Bacteriol 153: 458–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Gillham NW, Boynton JE (1985) Processing of the precursor to a chloroplast ribosomal protein made in the cytosol occurs in two steps, one of which depends on a protein made in the chloroplast. Mol Cell Biol 5: 1093–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G, Knight S, Andersson I, Branden CI, Lindqvist Y, Lundqvist T (1990. a) Comparison of the crystal structures of L2 and L8S8 Rubisco suggests a functional role for the small subunit. EMBO J 9: 2045–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G, Lindqvist Y, Brandedn C-I, Lorimer G (1986) Three dimensional structure of ribulose-1,5-bisphosphate carboxylase/oxygenase from Rhodospirillum rubrum at 2.9 A resolution. EMBO J 5: 3409–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G, Lindqvist Y, Lundqvist T (1990. b) Crystallographic refinement and structure of ribulose-1,5-bisphosphate carboxylase from Rhodospirillum rubrum at 1.7 A resolution. J Mol Biol 211: 989–1008 [DOI] [PubMed] [Google Scholar]
- Shapira M, Lers A, Heifetz P, Yrihimovitz V, Osmond BC, Gillham NW, Boynton JE (1997) Differential regulation of chloroplast gene expression in Chlamydomonas reinhardtii during photoacclimation: light stress suppresses synthesis of the Rubisco LSU protein while enhancing synthesis of the PSII D1 protein. Plant Mol Biol 33: 1001–1011 [DOI] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G (1997) RNA-binding proteins as regulators of gene expression. Curr Opin Genet Dev 7: 345–353 [DOI] [PubMed] [Google Scholar]
- Spreitzer R (1999) Questions about the complexity of chloroplast ribulose-1.5-biphosphate carboxylase/oxygenase. Photosynth Res 60: 29–42 [Google Scholar]
- Spreitzer RJ (1993) Genetic dissection of Rubisco structure and function. Annu Rev Plant Physiol Plant Mol Biol 44: 1–49 [Google Scholar]
- Spreitzer RJ (2003) Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch Biochem Biophys 414: 141–149 [DOI] [PubMed] [Google Scholar]
- Spreitzer RJ, Thow G, Zhu G (1995) Pseudoreversion substitution at large-subunit residue 54 influences the CO2/O2 specificity of chloroplast ribulose-bisphosphate carboxylase/oxygenase. Plant Physiol 109: 681–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara H, Yamamoto H, Shibata N, Inoue T, Okada S, Miyake C, Yokota A, Kai Y (1999) Crystal structure of carboxylase reaction-oriented ribulose 1, 5-bisphosphate carboxylase/oxygenase from a thermophilic red alga, Galdieria partita. J Biol Chem 274: 15655–15661 [DOI] [PubMed] [Google Scholar]
- Tabita FR (1999) Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth Res 60: 1–28 [Google Scholar]
- Tabita FR, McFadden BA (1974) D-Ribulose 1,5 diphosphate carboxylase from Rhodospirillum rubrum. I. Levels, purification and effects of metallic ions. J Biol Chem 249: 3453–3458 [PubMed] [Google Scholar]
- Taylor TC, Andersson I (1996) Structural transitions during activation and ligand binding in hexadecameric Rubisco inferred from the crystal structure of the activated unliganded spinach enzyme. Nat Struct Biol 3: 95–101 [DOI] [PubMed] [Google Scholar]
- Taylor TC, Backlund A, Bjorhall K, Spreitzer RJ, Andersson I (2001) First crystal structure of Rubisco from a green alga-Chlamydomonas reinhardtii. J Biol Chem 276: 48159–48164 [DOI] [PubMed] [Google Scholar]
- Thow G, Spreitzer RJ (1992) Missense mutations in the chloroplast rbcL gene that affect Rubisco holoenzyme assembly. In N Murata, ed, Research in Photosynthesis, Vol 3. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 633–636
- van der Vies SM, Bradley D, Gatenby AA (1986) Assembly of cyanobacterial and higher-plant ribulose-bisphosphate carboxylase subunits into functional homologous and heterologous enzyme enzyme molecules in Escherichia-coli. EMBO J 5: 2439–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Andrews TJ (2001. a) Plastome-encoded bacterial ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) supports photosynthesis and growth in tobacco. Proc Natl Acad Sci USA 98: 14738–14743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Andrews TJ (2001. b) The gene for the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) small subunit relocated to the plastid genome of tobacco directs the synthesis of small subunits that assemble into Rubisco. Plant Cell 13: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K, Girard-Bascou J, Wollman FA, Choquet Y (2004) Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J 23: 2696–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Irihimovitch V, Knopf JA, Cohen I, Orr-Dahan I, Nahum E, Keasar C, Shapira M (2004) RNA binding activity of the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit from Chlamydomonas reinhardtii. J Biol Chem 279: 10148–10156 [DOI] [PubMed] [Google Scholar]
- Youssefian S, Nakamura M, Orudgev E, Kondo N (2001) Increased cysteine biosynthesis capacity of transgenic tobacco overexpressing an O-acetylserine (thiol) lyase modifies plant responses to oxidative stress. Plant Physiol 126: 1001–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]