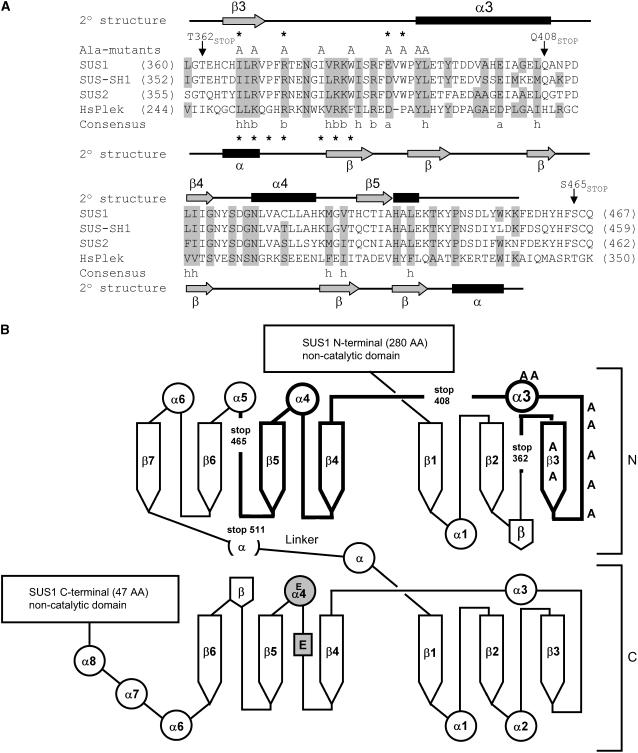
Figure 1.
SUS proteins can be modeled onto a GT-B-type glucosyltransferase fold and contain a region with sequence similarity to a PH domain. A, Alignment of amino acid sequences of the maize SUS isoforms SUS1 (P49036), SUS-SH1 (P04712), and SUS2 (Q8L5H0), with the C-terminal PH domain of human pleckstrin (HsPlek; P08567) by CLUSTALW (v 1.82). Secondary structure elements for SUS1 identified by the Jpred method are shown above the sequences, and those of pleckstrin below the sequences. Conservation of similar amino acids between HsPlek and the SUS isoforms is indicated by gray shading. Consensus residues are conserved across all four proteins. h, Hydrophobic; a, acidic; b, basic residues. The locations of Ala-substituted mutants are indicated by “A” and truncation (stop) mutants with arrows above the SUS1 sequence, residues identified in this study as important for membrane/lipid binding are marked with asterisks. Asterisks below the HsPlek sequence are important for PH domain membrane binding (Isakoff et al., 1998; Edlich et al., 2005). B, Diagram of SUS1 sequence showing Jpred secondary structural elements (α helices as circles, β strands as arrows) for the putative catalytic domain on a GT-B-type fold (drawn after MacGregor, 2002). Bold lines represent the PH-like domain of SUS1, locations of truncation mutants are indicated by “stop”, and Ala-substituted mutants by an “A”. The putative catalytic Glu residues are shown in gray.