Abstract
Phosphoprotein-binding domains are found in many different proteins and specify protein-protein interactions critical for signal transduction pathways. Forkhead-associated (FHA) domains bind phosphothreonine and control many aspects of cell proliferation in yeast (Saccharomyces cerevisiae) and animal cells. The Arabidopsis (Arabidopsis thaliana) protein kinase-associated protein phosphatase includes a FHA domain that mediates interactions with receptor-like kinases, which in turn regulate a variety of signaling pathways involved in plant growth and pathogen responses. Screens for insertional mutations in other Arabidopsis FHA domain-containing genes identified a mutant with pleiotropic defects. dawdle (ddl) plants are developmentally delayed, produce defective roots, shoots, and flowers, and have reduced seed set. DDL is expressed in the root and shoot meristems and the reduced size of the root apical meristem in ddl plants suggests a role early in organ development.
During development, protein-protein interactions play a critical role in assembling signaling complexes that are essential for cells to grow and differentiate properly in response to neighboring cells and their environment. These protein-protein interactions are often mediated by modular protein domains, which bind specific motifs on target proteins. Protein-binding domains that recognize phosphorylated binding targets are a prevalent mechanism involved in mediating interactions between proteins. Several classes of phosphobinding domains have been identified and classified by their ability to target specific motifs. A well-characterized phosphobinding domain is the Src homology-2 domain. Src homology-2 domains bind a phospho-Tyr containing protein targets and are involved in the regulation of pathways that include cellular functions such as adhesion and hormone responses (Pawson, 2003). Other phospho-Tyr-binding domains have been described, as well as domains that bind phospho-Thr and phospho-Ser (Yaffe and Smerdon, 2001). For example, WW domains and 14-3-3 domains bind proteins with motifs that contain phospho-Ser to regulate cell cycle transitions (Yaffe and Smerdon, 2001; Pawson, 2003).
Another large family of protein domains, the forkhead-associated (FHA) domains, mediate protein-protein interactions by targeting motifs that contain phospho-Thr (Durocher et al., 1999). First identified in forkhead transcription factor complexes (Hofmann and Bucher, 1995), the structure and function of FHA domains from several different proteins have been determined (Li et al., 2000; Durocher and Jackson, 2002). Three-dimensional structures of FHA domains solved by NMR and x-ray crystallography show that FHA domains take a β-sheet sandwich formation (Liao et al., 1999, 2000; Stavridi et al., 2002; Lee et al., 2003; Li et al., 2004). Strands of the sheets are connected by mobile loops, where conserved residues are located. Conserved residues localize to one surface of the folded protein, assembling the site required for interaction with phosphorylated targets (Durocher et al., 2000). Variable loop residues surrounding the conserved residues not only help establish interactions between conserved residues and their binding partners, but also stabilize the overall protein structure (Yongkiettrakul et al., 2004). These phosporylation-dependent interactions are now known to play a role in many processes critical for cell proliferation, such as DNA damage response and repair and mitotic progression (Sun et al., 1998; Scolnick and Halazonetis, 2000).
In plants, receptor-like kinases (RLKs) regulate a variety of signaling pathways involved in regulating processes such as plant growth and pathogen responses (Williams et al., 1997; Stone et al., 1998; Gomez-Gomez et al., 2001). In the search for RLK signaling pathway components, interaction cloning identified kinase-associated protein phosphatase (KAPP) as a binding partner of HAESA (Stone et al., 1994). KAPP is a type 2C protein phosphatase that contains a FHA domain as well as a type I signal anchor that directs the protein to the plasma membrane. In addition, a KAPP homolog was identified in maize (Zea mays) and also interacts with several RLKs (Braun et al., 1997). Using the Arabidopsis (Arabidopsis thaliana) protein, it was determined that a 119-amino acid stretch of KAPP containing the FHA domain is required for phosphorylation-dependent interactions with RLKs (Li et al., 1999). While several of the FHA domains discovered in humans and yeast (Saccharomyces cerevisiae) have been characterized, KAPP remains the only well-studied plant FHA domain gene. The FHA domain of KAPP recognizes a wide variety of phosphorylated RLKs at the cell membrane. KAPP does not interact with all RLKs, but has been shown to interact with CLAVATA1 (CLV1), HAESA, FLAGELLIN-SENSITIVE 2, KI DOMAIN INTERACTING KINASE 1, CELL WALL-ASSOCIATED KINASE 1, and SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1 (Stone et al., 1994, 1998; Braun et al., 1997; Williams et al., 1997; Gomez-Gomez et al., 2001; Park et al., 2001; Shah et al., 2002). These RLKs are a small portion of the 400 plus genes in the RLK family, but represent the wide array of signaling pathways potentially regulated by KAPP. Interaction with the RLK most likely brings the type 2C protein phosphatase domain of KAPP in proximity with the kinase domain of the RLK, negatively regulating the kinase's activity through dephosphorylation.
There are 15 genes in the Arabidopsis genome that encode predicted proteins that contain a FHA domain. Only two of these genes, KAPP and ABA1 (an abscisic acid-insensitive mutant), have been studied in detail (Stone et al., 1994; Xiong et al., 2002). ABA1 is a zeazanthin epoxidase that functions in the abscisic acid biosynthesis pathway. ABA1 contains a predicted FHA domain, but the function is yet to be determined. Plants lacking ABA1, and therefore abscisic acid, have many altered responses and developmental defects. Mutant plants are sensitive to osmotic stress and drought and have a reduced hydrotropic response (Takahashi et al., 2002; Xiong et al., 2002). Two Arabidopsis genes (At3g07220 and At3g07260) are homologous to NtFHA1, a tobacco (Nicotiana tabacum) gene whose FHA domain is capable of substituting for a yeast FHA domain in transcription activities (Kim et al., 2002); however, its function in planta is unknown.
The FHA domain is associated with a variety of proteins with differing functions. We are interested in investigating the role of the other FHA domain-containing proteins in Arabidopsis to determine the functions of these genes in plants. Screening T-DNA populations for insertions in several of the FHA domain-containing genes identified a loss-of-function mutant that was developmentally delayed, had defective vegetative and reproductive organs, and severely reduced seed set.
RESULTS
Most of the Arabidopsis genes that are predicted to contain a FHA domain share little homology with each other, or with other proteins. Thus, predicting the function of these FHA domain genes based on sequence similarity is difficult, even with some knowledge about the function of KAPP and ABA1. Therefore, we took a functional genomics approach to determine whether any other FHA domain genes play an important role in plant growth and development. To isolate loss-of-function mutants, we screened several T-DNA populations. We obtained 38 lines from three T-DNA populations: four Wisconsin lines, 24 SAIL lines, and 10 SALK lines (Sussman et al., 2000; Sessions et al., 2002; Alonso et al., 2003). All together, 13 genes were represented in these 38 lines (Supplemental Table I). Plants were screened for developmental phenotypes due to gene disruption by the T-DNA.
One loss-of-function mutant identified in our screens displayed pleiotropic effects. Mutant plants exhibit shortened roots, delayed flowering time, altered floral organ numbers, defective floral organs, and reduced fertility. Because of the prolonged growth period of the mutant, the corresponding gene was named DAWDLE (DDL).
ddl Alleles
T-DNA insertion in DDL (At3g20550) results in an altered development. The DDL gene consists of 10 exons, with the FHA domain spanning exons six through nine. The deduced DDL protein is 314 amino acids and includes several putative nuclear localization sequences at the N terminus, and a FHA domain spanning residues 218 to 282 near the C terminus (Fig. 1B). No other specific domains or features could be identified besides the FHA domain. In addition, there is no apparent homology to any other characterized proteins.
Figure 1.
Identification and complementation of ddl alleles. A, Ecotype Wassilewskija of Arabidopsis (WS), ddl-1 complemented with genomic DDL, and the ddl-1 mutant. B, DDL gene and protein structure. ddl-1 and ddl-2 are T-DNA insertion alleles and ddl-3 is a TILLING point mutation. Three nuclear localization signals are predicted at the N terminus of the protein. The FHA domain stretches from residues 218 to 282 at the C terminus of the protein. C, RT-PCR results show that DDL RNA cannot be amplified in ddl-1 or ddl-2 homozygous mutants. Labels above gels indicate RNA source. Lines below indicate primers used. CLV1 primers were used as RNA quality controls.
Two independent ddl alleles were identified from the Wisconsin T-DNA populations. Both display the same phenotype and segregate 3:1 wild type:mutant from a heterozygous self pollination (23.9% homozygous mutants). This suggests that the ddl phenotype is due to a recessive sporophytic mutation caused by the T-DNA insertion. To prove that the mutant phenotype is caused by the insertion of the T-DNA in DDL, we complemented the mutant phenotype with a wild-type copy of the DDL gene. Transformation of ddl-1 homozygous plants with the transgene containing the genomic DDL sequence complements all of the mutant phenotypes (Fig. 1A). The genomic sequence of DDL, including 1,300 bp upstream and downstream of the coding region, was transformed into ddl-1 plants. Complemented plants had the same low level of expression seen in wild-type plants (data not shown). Therefore, loss of function of ddl appears to be the cause of the mutant phenotypes observed in these T-DNA lines.
To verify whether the two ddl alleles are null alleles, we used both northern blot and reverse transcription (RT)-PCR to detect DDL transcripts in the mutant alleles. No DDL transcript is detectable in ddl-1 or ddl-2 plants by northern blot (data not shown). This result was confirmed by RT-PCR: While the DDL transcript can be amplified from total RNA from wild-type plants, it cannot be amplified from ddl-1 or ddl-2 RNA (Fig. 1C). Together these data suggest the two ddl alleles are null alleles.
To determine the insertion site of the T-DNA in the DDL gene, a T-DNA left border primer and a DDL-specific primer were used to amplify and sequence the flanking genomic sequence. ddl-1 contains a single insertion at 1,112 bp, near the intron 3/exon 4 border, while the T-DNA insertion in ddl-2 is 25 bp upstream of the predicted initiation codon.
ddl Mutations Cause Pleiotropic Growth Defects
During the vegetative stage, ddl plants are easily identified by their small size. The rosettes are smaller than wild type, the leaves are narrower with a pointed shape and become curly as the plants grow (Fig. 1A). When grown in soil for 31 d, rosettes of wild-type plants are 5.9 cm (±1.23 sd) across at the widest point, while rosettes of ddl-1 and ddl-2 mutants are 2.8 cm (±0.59 sd) and 2.6 cm (±0.69 sd) wide, respectively. Under standard growth conditions, ddl plants are short and bushy, producing many fruit. Under these conditions the mutant plants stay green and continue to grow long after wild-type plants of the same age have set seeds and senesced. If the ddl plants are stressed (drought, over watering, high temperature, or herbivory), they display an increased stress phenotype compared to wild-type plants, resulting in a very short and single inflorescence stem that produces few fruit.
ddl Flowers Have Reduced Numbers of Floral Organs
ddl flowers exhibit defective floral organs as well as altered numbers of floral organs (Fig. 2; Table I). While there was no evidence of organ conversion, the mutant flowers had significantly fewer floral organs than wild type in the outer three whorls. To assess the changes in floral organ number, offspring from a self-fertilized heterozygous plant for ddl-1 were genotyped to identify individuals heterozygous or homozygous for ddl-1, and wild-type plants lacking the insertion. In ddl-1 homozygous mutants, the number of sepals was reduced and some sepals appeared to be fused together. The petals of mutant flowers appear almost shriveled and are smaller than wild type. The ddl-1 flowers also had altered petal numbers; over half of ddl-1 flowers (53%) had fewer than four petals. A small percentage of flowers had an extra petal or sepal. Wild-type plants showed only minimal variation in the first two whorls.
Figure 2.
ddl phenotypes. In all sections wild-type plants are on the left and ddl-1 plants are on the right. A, Inflorescences. B, Flowers. C, Fruit.
Table I.
Floral organ numbers
Total indicates number of flowers analyzed per genotype. WS, Ecotype Wassilewskija of Arabidopsis.
| Organ | Number of Organs per Flower
|
Total | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| Sepals | ||||||
| WS | 0 | 0 | 3 | 197 | 0 | 200 |
| ddl-1 | 0 | 16 | 84 | 88 | 0 | 181 |
| ddl-2 | 0 | 0 | 8 | 56 | 6 | 70 |
| Petals | ||||||
| WS | 0 | 0 | 3 | 197 | 0 | 200 |
| ddl-1 | 1 | 17 | 78 | 79 | 6 | 181 |
| ddl-2 | 1 | 0 | 12 | 51 | 6 | 70 |
| Stamens | ||||||
| WS | 0 | 1 | 43 | 85 | 71 | 200 |
| ddl-1 | 1 | 42 | 113 | 25 | 0 | 181 |
| ddl-2 | 2 | 10 | 42 | 15 | 1 | 70 |
The number of stamen per flower varied in wild-type controls from 35.5% containing six stamens, while 42.5% had five stamens, and 22% had fewer than five. Yet of the 181 ddl-1 flowers observed, none contained six stamens. Instead, the majority (62.4%) of ddl-1 flowers had only four stamens. Anther morphology was also altered. When compared to wild type, the ddl anthers appear collapsed, although they produced pollen that is viable, as determined by staining with Alexander solution (Alexander, 1969; data not shown).
Reduced Fertility in ddl Mutants
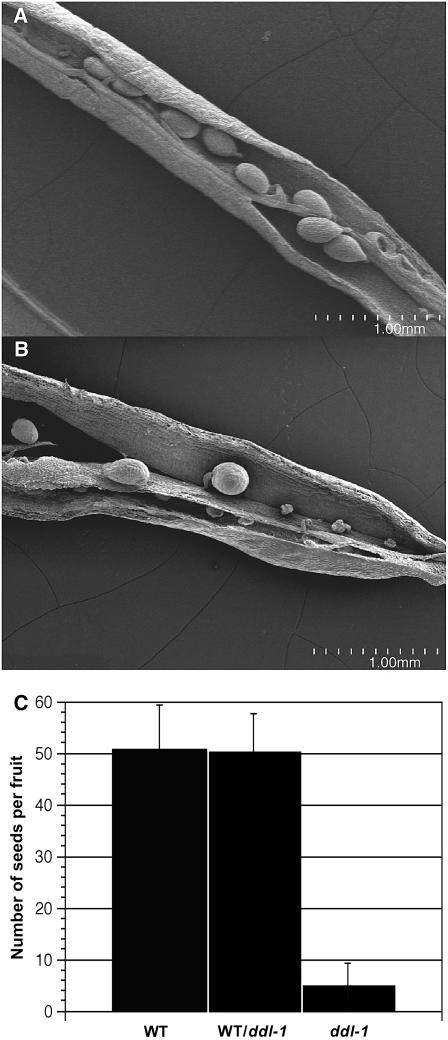
The most dramatic phenotype due to loss of ddl function is reduced fertility. In optimal growth conditions, ddl plants produce many fruit but set very few seeds per silique. The siliques are abnormally shaped and smaller than wild-type fruit (Fig. 2). To determine the number of seeds produced in each silique, offspring from a self-pollinated plant heterozygous for ddl-1 were genotyped to identify wild-type, heterozygous, and homozygous mutant plants. Wild-type plants set 50 seeds (±8.59 sd) per silique while heterozygous plants set 50.4 seeds (±7.34 sd) per silique. In contrast, ddl-1 fruit set an average of 4.6 seeds (±4.24 sd) per silique. As seen with scanning electron microscopy (SEM), many ovule primordia are initiated in the mutants, but few develop into seeds (Fig. 3). Furthermore, pollinating ddl stigma with wild-type pollen did not restore seed production. In 73 crosses only 3.4 seeds per silique (±2.95 sd) developed. This observation suggests the loss of DDL function in the sporophytic tissue of ddl siliques compromises seed development. The outcross with wild-type pollen also reinforces the segregation data and supports the conclusion that the ddl defect is sporophytic.
Figure 3.
Seed set in ddl mutants is decreased. A, SEM images from wild-type silique. B, ddl-1 silique. C, Graph showing average number of seeds per fruit. Error bar = sd. Sample numbers: wild-type (WT) fruit (n = 202); WT/ddl-1 fruit (n = 150); and ddl-1 fruit (n = 196).
Root Growth Is Inhibited in ddl Plants Due to Reduced Meristem Size
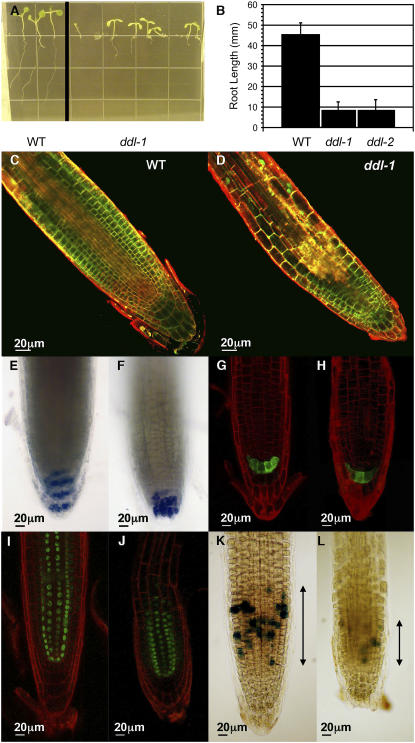
ddl roots have altered growth as well. When grown on vertical plates, 100% of ddl seeds germinated, but as many as 23% had severe problems with root growth after germination. Many roots grew in the wrong direction, either up or out of the plate. Other roots never grew more than 2 mm after more than 10 d in the light (data not shown). In addition, the root length is affected in the mutant. After 10 d, roots of wild-type plants grown on vertical plates were 45.2 mm (±5.79 sd) long. ddl-1 and ddl-2 roots are shorter, growing to only 8.2 mm (±4.26 sd) and 8.1 mm (±5.27 sd) long, respectively (Fig. 4, A and B). In an effort to understand the growth defects in roots of ddl mutants, confocal microscopy was used to visualize the cells of growing ddl roots. Staining of the plasma membrane with FM-143 and Syto 24 shows that in ddl mutants the elongation zone is closer to the root tip than in wild-type plants (Fig. 4, C and D). Therefore, the root morphology is changed in the mutant. To test whether cell identity and fate are affected in the ddl mutant, we stained the starch granules in the amyloplast and examined the expression of the enhancer trap line Q1630 and the SHORT-ROOT (SHR) green fluorescent protein (GFP) fusion protein. Both the starch granules and Q1630 are normally excluded from the columella cell initials and present only in mature columella cells (Sabatini et al., 2003), while SHR is expressed in the stele tissue (Helariutta et al., 2000). For these three markers, no patterning differences could be seen between the wild type and the ddl mutant (Fig. 4, E–J). Therefore, cell fate and patterning of the root appear not to be affected in the ddl mutant. To test whether cell size and/or cell number is affected in the ddl mutant, we measured the cell size and counted the cell number in roots of both wild type and mutant. We measured the size of 30 cells from six wild-type and mutant plants each. The size of fully elongated cells is 2,266 μm2 (±603 sd) for the wild type and 2,234 μm2 (±543 sd) for the ddl mutant. Thus, there is no difference in cell size between the wild type and the ddl mutant. To test whether cell number is affected in the mutant, we counted the number of cells and measured the length between the root tip and the first cell that initiates a root hair. For the wild type, the length between the root tip and the first cell that initiates a root hair is 977 μm (±110 sd) and the number of cells is 44 (±2.3 sd). For the mutant, the length is 545 μm (±47 sd) and the number of cells is 27.2 (±4.2 sd). Therefore, the cell number is reduced in the mutant resulting in shorter roots.
Figure 4.
Root growth and morphology of ddl mutants. A, Seeds were germinated on vertical Murashige and Skoog plates and grown for 10 d. B, Graph showing average root length for wild-type, ddl-1, and ddl-2 plants. Error bars show sd. Sample numbers: wild type (WT) = 91, ddl-1 = 66, and ddl-2 = 68. C and D, Confocal images of root tips. E and F, Starch granule staining. G and H, Enhancer trap line Q1630. I and J, pSHR::GFP-SHR. K and L, Marker line FA4C cyclin B1;1 GUS. Plasma membranes were stained with Syto24 and FM-143 (C and D) and propidium iodide (G to J). E, G, I, and K are wild type. F, H, J, and L are ddl-1.
To better understand the reduction of cell number in the ddl mutant, we used a marker for the G2/M phase of the cell cycle. The FA4C marker line is a cyclin B1;1 β-glucuronidase (GUS) line with a dead box (Colon-Carmona et al., 1999). In the wild type, the number of GUS-positive cells is 28.5 (±4.4 sd; 15 plants), and 11 cells (±3.8 sd) in the mutant (nine plants). Therefore, the number of cell divisions is reduced in the mutant. To test whether the reduced number of cell divisions is due to a reduced meristematic zone in the mutant, we measured the length of the meristematic zone in the root that contains GUS-positive cells. The length of the cell division zone of the root that contains GUS-positive cells is 139 μm (±12.8 sd) for the wild type and 78.8 μm (±23.4 sd) for the mutant. These results suggest the mutant phenotype is due to a perturbation in cell divisions, not a change in cell size or cell fate.
DDL Expression Pattern and Localization
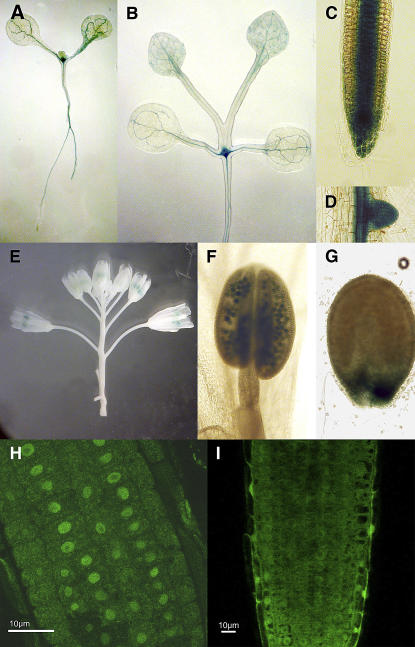
The GUS gene was expressed under the control of the DDL promoter (Fig. 5, A–G). We used the 1,300 bp upstream of the DDL gene, which was sufficient to rescue the ddl mutant. DDL is expressed in the root and vegetative meristems, but no expression can be detected in the inflorescence and floral meristems. In addition, GUS can be detected in the lateral roots and the vascular strands of roots and leaves. For the reproductive organs, DDL is expressed in both pollen and developing seeds.
Figure 5.
DDL::GUS expression pattern and localization of DDL-GFP protein. A to G, DDL::GUS. A, 2-d-old seedling. B, 5-d-old seedling. C, Root tip. D, Lateral root. E, Inflorescence. F, Anther. G, Seed. H, 35S::GFP expressed in wild-type (WT) plants is cytoplasmic. I, DDL-GFP expressed behind a 35S promoter in WT plants localizes to the nucleus. Images show root tips of each plant.
Several nuclear localization sequences are predicted in the N-terminal part of DDL. To test whether DDL localizes to the nucleus, DDL was fused to the GFP. The DDL-GFP C-terminal fusion was expressed under the 35S promoter and transformed into wild-type plants. As a control, plants were also transformed with a 35S::GFP construct. Roots of transformed plants were visualized with a confocal microscope (Fig. 5, H and I). The florescence from the GFP protein localizes in the nucleus of the root tip cells in 35S::DDL-GFP plants, whereas the control 35S::GFP construct associates with the periphery of the nucleus. Multiple lines from independent transformation events displayed similar expression patterns. The nuclear localization of DDL-GFP supports the possibility that DDL may function in the nucleus.
DISCUSSION
Two T-DNA insertion lines that interrupt At3g20550 were identified. The corresponding gene was named DDL because of slow, prolonged growth of the mutant plants. Each of these T-DNA alleles has no detectable DDL mRNA, and complementation with genomic DDL sequence restores the mutant phenotypes to wild type, confirming that loss of DDL function is the cause of the mutant phenotypes. In addition, we identified ddl-3, a TILLING (Till et al., 2003) allele that changes one of the conserved residues of the FHA domain, that displays the same phenotypes as the null alleles and suggests the FHA domain is required for DDL function.
In ddl-1 and ddl-2 plants, changes in morphology can be seen in the first true leaves, as they are small and pointed. As the plant grows, the leaves begin to curl, and the mature rosette is much smaller than wild type. ddl flowers are also dramatically affected. The most immediate difference is that the petals never expand and emerge from the bud. The petals are small, sometimes appearing filamentous, and wilted. This may be the reason the fruit protrude out of the bud earlier than wild type. The fruit are often short and curved as they develop. Because DDL is expressed in meristematic tissues it is likely that DDL has a function early in the development of these organs. Stamen development does appear to be defective, but segregation of offspring from a self-pollinated heterozygous plant and staining of viable pollen with Alexander solution show that ddl pollen is viable.
Upon further inspection, the floral organs are not only different in appearance, but also fewer in number. The sepal number is variable and sometimes they appear to be fused. The number of petals is decreased in the majority of ddl flowers, and the most significant difference is in the third whorl. All but one ddl flower observed lacked at least one stamen, and more than 86% of ddl-1 flowers contained four or fewer. Although the ddl ovary contains two fused carpels like wild type, and many ovules initially develop, very few develop into seeds. Seed set was reduced by more than 90%.
Root growth is also abnormal in ddl mutants. ddl mutants have smaller roots due to an abridged root meristematic zone, resulting in a reduction of cell divisions and cell number. The ddl growth defects and localization to the nucleus supports the idea that DDL may have a role in the regulation of cell proliferation. It is interesting to note that most of FHA domain-containing genes that have been characterized in other organisms localize to the nucleus and are involved in some aspect of cell proliferation. Some FHA domain-containing proteins, like Rad53 in yeast and Chk2 in humans, respond to DNA damage (Sun et al., 1998; Bell et al., 1999). When these genes are mutated, cell cycle checkpoints are compromised and often cause cancer or other diseases in humans (Varon et al., 1998; Bell et al., 1999; Chehab et al., 2000; Hirao et al., 2000). Another group of FHA domain-containing genes control mRNA splicing during checkpoints (Boudrez et al., 2000, 2002; Vulsteke et al., 2004). FHA domains of other proteins functions in mitotic chromosome movement (Scolnick and Halazonetis, 2000). A few yeast transcription factors use their FHA domains to facilitate formation of active transcription complexes (Loy et al., 1999; Koranda et al., 2000). To our knowledge, DDL is the first example of a nuclear, localized FHA domain-containing protein functioning in plants, and may also have a role in cell division or differentiation.
Although we do not know the direct mechanism of DDL, our data showing expression of DDL in the meristem, and decreased meristem size in ddl roots suggest that it plays a role in the early stages of organ development. A mutation that results in a decreased number of cells in the root tips might also affect cell proliferation in other meristematic regions. If cell proliferation is compromised in ddl plants and cellular material used for new organ production is limited, then the dramatic shoot and root phenotypes of ddl mutants are not unanticipated. Plants lacking CURLY LEAF (CLF) gene function are defective in cell proliferation and elongation during leaf development (Kim et al., 1998). The resulting smaller, misshapen, curly clf leaves are similar to ddl rosette leaves. CLV1 is another gene that controls meristematic cell number. clv1 mutants have enlarged shoot apical meristems (SAMs) with an increased number of undifferentiated cells, supporting the growth of extra floral organs (Leyser and Furner, 1992; Clark et al., 1997). In contrast, the lower number of SAM cells in plants lacking the WUS gene have smaller meristems and fewer auxiliary meristems due to failure in stem cell maintenance (Laux et al., 1996; Mayer et al., 1998). Another example of extremely reduced meristem size is SHOOT MERISTEMLESS (STM). STM maintains the undifferentiated state of meristematic cells and without it no SAM develops (Long et al., 1996). All of these mutants show that changes in organ number (decrease or increase) can be caused by changes in meristematic cell number. If DDL does promote cell proliferation in meristematic regions, the hindered root growth and lack of floral organs could be a direct effect of lowered cell numbers.
Many of the other FHA domain genes that have been characterized function in the nucleus, and it is not unfounded to suggest DDL has a nuclear function as well. DDL includes putative nuclear localization signals, and GFP localization suggests nuclear localization, which would be consistent with a roll in cell cycle or cell proliferation events. There are many possible roles that DDL might play in cell division; it could monitor DNA stability like Rad53 and regulate cell cycle events, or DDL could influence transcription activation by recruiting proteins to transcription complexes. The pleiotropic phenotype suggests that if DDL does influence gene expression, the affected genes are involved in early organ development. Future experiments that characterize interaction partners or identify changes in gene expression patterns in ddl mutants will help uncover the role of DDL in plant development.
MATERIALS AND METHODS
Mutant Identification
T-DNA insertion lines were obtained from the University of Wisconsin, SALK, and Syngenta (Sussman et al., 2000; Sessions et al., 2002). Plants were grown on soil either in a greenhouse or growth chamber kept at 21°C in 16-/8-h light/dark cycles. Growth chambers were kept at 60% humidity. For genotyping, a 2 cm3 piece of leaf tissue was collected and homogenized in 100 mm Tris, pH 8, 50 mm EDTA, 500 mm NaCl, 10 mm β-mercaptoethanol, and 1.25% SDS. DNA was extracted by using 3 m potassium acetate and 5 m acetic acid, and DNA was precipitated with isopropanol.
A T-DNA left border primer and gene-specific primer located approximately 500 bp outside of the coding region were used to amplify the region around the insertion. The following T-DNA left border primers were used: Wisconsin lines, J270 (5′-TTTCTCCATATTGACCATCATACTCATTG-3′), and the following primers were used as gene-specific primers: AT3g20550 (5′-TCACTTGCTTCCCCATCATACCATCTGGT-3′ and 5′-TATGACCTAAGAGGCTACGTATGTTCGCT-3′). To determine the site of the T-DNA insertion, PCR products were sequenced using the ABI Prism d-Rhodamine sequencing kit (part No. 403042), and the T-DNA primer was used as the sequencing primer. An ABI3700 sequencer was used for sequencing, and DNAStar software was used to analyze the results.
Southern-Blot Analysis
Genomic DNA was isolated from a heterozygous individual for ddl using the Nucleon Phytopure kit (Amersham) and purified using the QbioGene GenecleanTurbo kit (Product no. 1102-600). DNA was digested using BamH1, EcoRI, KpnI, and SacI and then run on an agarose gel and transferred to nylon membrane. The membrane was dried and hybridized with a T-DNA-specific probe. Primers used to amplify the probe were 5′-GTAATACGACTCACTATAGGGCGAATTGGAGCT-3′ and 5′-GGAACGTCTTCTTTTTCCACGATGTTCCTCGTG-3′.
RNA Isolation and Analysis
Total RNA was extracted using the RNeasy kit (Qiagen #75144). For the RT-PCR reactions, the Invitrogen SuperScript One-Step RT-PCR kit (#10928-034) was used with the following primers specific to the DDL cDNA: 5′-TCTAAGCTTATGGCTCCTAGTTCTAGGTCTC-3′ and 5′-TCTAAGCTTTCACTCGGCAGAATTCTCGTGC-3′.
For northern blots, 20 μg of RNA were run on an RNA gel, transferred to a membrane, and probed with the DDL cDNA sequence.
Complementation
The genomic sequence of DDL was amplified using primers with XmaI restriction sites on the 5′ ends for cloning into expression vectors (primers: G20550XmaI-5′, 5′-TCTACCCGGGTGACCTAATTTGGACTTTGTCATAG-3′, G20550XmaI-3′, and 5′-TCTACCCGGGTCATGATGAAGTAATCTTGGAAGGA-3′). DDL was cloned into the pGEM T-easy vector (Promega #TM042), then into pCAMBIA2300 (CAMBIA) using XmaI (p2300GDDL). Flowers of homozygous ddl-1 plants were dipped twice in GV3101 Agrobacterium tumefaciens cells that had been transformed with p2300GDDL, allowing a week of growth between dips (Clough and Bent, 1998). Transformed seeds were selected on 50 μg/mL kanamycin plates, then transplanted to soil.
Growth Measurements
Rosette width was measured across the widest part while in the soil. Floral organs were counted under a dissecting scope, while a light microscope was used for seed counting. To measure root growth, seeds were plated on vertical one-half Murashige and Skoog plates, vernalized at 4°C for 48 h, and then moved into light. Root measurements were either made between the root tip and the point of root-shoot transition to measure the overall root length, or between the root tip and the first cell that initiates a root hair. The number of cells was counted under a light microscope. The size of cells above the first cell that initiates a root hair was measured using the ImageJ software (http://rsb.info.nih.gov/ij/). For the FA4C cycline line, the number of GUS-positive cells and the length of the zone that contains GUS-positive cells were measure under a light microscope.
Microscopy
Tissues were collected from wild-type and ddl plants and fixed for SEM using the methods outlined in Lease et al. (2001). Images were collected with a Hitachi S4700 cold-cathode, field-emission-scanning electron microscope.
To obtain images of root structure, plants were grown vertically on one-half Murashige and Skoog plates for 5 d in yellow light. A total of 500 nm Syto24 (Molecular Probes #S-7559) was dropped onto the plates where the roots were growing and incubated at room temperature for 45 min. Then 50 μg/mL FM143 (Molecular Probes #T-3163) was dropped onto the roots and left at room temperature for 30 min. Plates were incubated in the dark to prevent dye bleaching. Images of root structure, DDL-GPF, Q1630, and pSHR::GFP-SHR lines were collected with a Bio-Rad Radiance 2000 coupled with an Olypmus IX70 inverted microscope. For Q1630 and pSHR::GFP-SHR lines, the plasma membrane of the root was stained with propidium iodide. Starch granules were visualized as described (Willemsen et al., 1998).
GFP Fusions
The DDL cDNA was amplified from wild-type plants (Columbia ecotype) by RT-PCR, then reamplified with primers containing restriction sites for cloning into the SK+ plasmid containing the GFP sequence (primers: 20550cDNAKpnI-5′, 5′-TCTGGTACCATGGCTCCTAGTTCTAGGTCTCCTTC-3′, 20550cDNABamHI-3′, and 5′-TCTGGATCCCTCGGCAGAATTCTCGTGCAACAGAC-3′). The cDNA was then ligated into the GFP plasmid. The DDL-GFP fusion was amplified for cloning into the pKAN plasmid for transformation (Li et al., 2002; primers: 20550cDNAKpnI-5′, 5′-TCTGGTACCATGGCTCCTAGTTCTAGGTCTCCTTC-3′, T3-KpnI-3′GFP, and 5′-TCTGGTACCATTACCCCTCACTAAAGGGAAGA-3′). DDL-GFP pKAN was transformed into GV3101 cells for floral dip (Clough and Bent, 1998). Transformed seeds were selected on 50 μg/mL KAN plates and transplanted to soil.
DDL Promoter-GUS Assay
The 1.3 kb sufficient to complement the ddl mutant was used as the DDL promoter. A primer pair was used to clone the DDL promoter from genomic DNA using PCR: 5′-TCTACCCGGGTGACCTAATTTGGACTTTGTCA-3′ and 5′-TCTACCCGGGGGAGTCAGGCACGTTTTCTCT-3′. Two SmaI sites at the end of the primer sequences were introduced restriction sites used for cloning. The PCR products were inserted into the promoterless GUS transformation vector pBIG-GUS and introduced into wild type via Agrobacterium GV-3101-mediated transformation as described before (Clough and Bent, 1998). Histochemical staining for GUS activity was performed with chromogenic substrate 5-bromo-4-chloro-3 indolyl glucuronide as detailed (Craig, 1992).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NM_112947 (DDL, At3g20550).
Supplementary Material
Acknowledgments
We thank Peter Doerner for the FA4C and Ben Sheres for the Q1630 and pSHR∷GFP-SHR seeds. We would like to thank the Molecular Cytology Core for their assistance in image collection. We are also grateful for the input and assistance provided by Jia Li, Jiangqi Wen, and Jason Doke at different stages of the project. We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis TDNA insertion mutants.
This work was supported by the National Science Foundation (grant no. MCB 0112278).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: John C. Walker (walkerj@missouri.edu).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.076893.
References
- Alexander MP (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, Lubratovich M, Verselis SJ, Isselbacher KJ, Fraumeni JF, et al (1999) Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science 286: 2528–2531 [DOI] [PubMed] [Google Scholar]
- Boudrez A, Beullens M, Groenen P, Van Eynde A, Vulsteke V, Jagiello I, Murray M, Krainer AR, Stalmans W, Bollen M (2000) NIPP1-mediated interaction of protein phosphatase-1 with CDC5L, a regulator of pre-mRNA splicing and mitotic entry. J Biol Chem 275: 25411–25417 [DOI] [PubMed] [Google Scholar]
- Boudrez A, Beullens M, Waelkens E, Stalmans W, Bollen M (2002) Phosphorylation-dependent interaction between the splicing factors SAP155 and NIPP1. J Biol Chem 277: 31834–31841 [DOI] [PubMed] [Google Scholar]
- Braun DM, Stone JM, Walker JC (1997) Interaction of the maize and Arabidopsis kinase interaction domains with a subset of receptor-like protein kinases: implications for transmembrane signaling in plants. Plant J 12: 83–95 [DOI] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Appel M, Halazonetis TD (2000) Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev 14: 278–288 [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colon-Carmona A, You R, Haimovitch-Gal T, Doerner P (1999) Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Craig S (1992) The GUS reporter gene. Application to light and transmission electron microscopy. In GSR Gallagher, ed, US Protocols. Using the GUS Gene as a Reporter of Gene Expression. Academic Press, New York, pp 115–124
- Durocher D, Henckel J, Fersht AR, Jackson SP (1999) The FHA domain is a modular phosphopeptide recognition motif. Mol Cell 4: 387–394 [DOI] [PubMed] [Google Scholar]
- Durocher D, Jackson SP (2002) The FHA domain. FEBS Lett 513: 58–66 [DOI] [PubMed] [Google Scholar]
- Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB (2000) The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell 6: 1169–1182 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Bauer Z, Boller T (2001) Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13: 1155–1163 [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW (2000) DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287: 1824–1827 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P (1995) The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem Sci 20: 347–349 [DOI] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H (1998) The CURLY LEAF gene controls both division and elongation of cells during the expansion of the leaf blade in Arabidopsis thaliana. Planta 206: 175–183 [DOI] [PubMed] [Google Scholar]
- Kim M, Ahn JW, Song K, Paek KH, Pai HS (2002) Forkhead-associated domains of the tobacco NtFHA1 transcription activator and the yeast Fhl1 forkhead transcription factor are functionally conserved. J Biol Chem 277: 38781–38790 [DOI] [PubMed] [Google Scholar]
- Koranda M, Schleiffer A, Endler L, Ammerer G (2000) Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature 406: 94–98 [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jurgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Lease KA, Wen J, Li J, Doke JT, Liscum E, Walker JC (2001) A mutant Arabidopsis heterotrimeric G-protein beta subunit affects leaf, flower, and fruit development. Plant Cell 13: 2631–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GI, Ding Z, Walker JC, Van Doren SR (2003) NMR structure of the forkhead-associated domain from the Arabidopsis receptor kinase-associated protein phosphatase. Proc Natl Acad Sci USA 100: 11261–11266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HM, Furner IJ (1992) Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development 116: 397–403 [Google Scholar]
- Li H, Byeon IJ, Ju Y, Tsai MD (2004) Structure of human Ki67 FHA domain and its binding to a phosphoprotein fragment from hNIFK reveal unique recognition sites and new views to the structural basis of FHA domain functions. J Mol Biol 335: 371–381 [DOI] [PubMed] [Google Scholar]
- Li J, Lee GI, Van Doren SR, Walker JC (2000) The FHA domain mediates phosphoprotein interactions. J Cell Sci 113: 4143–4149 [DOI] [PubMed] [Google Scholar]
- Li J, Smith GP, Walker JC (1999) Kinase interaction domain of kinase-associated protein phosphatase, a phosphoprotein-binding domain. Proc Natl Acad Sci USA 96: 7821–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Liao H, Byeon IJ, Tsai MD (1999) Structure and function of a new phosphopeptide-binding domain containing the FHA2 of Rad53. J Mol Biol 294: 1041–1049 [DOI] [PubMed] [Google Scholar]
- Liao H, Yuan C, Su MI, Yongkiettrakul S, Qin D, Li H, Byeon IJ, Pei D, Tsai MD (2000) Structure of the FHA1 domain of yeast Rad53 and identification of binding sites for both FHA1 and its target protein Rad9. J Mol Biol 304: 941–951 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Loy CJ, Lydall D, Surana U (1999) NDD1, a high-dosage suppressor of cdc28-1N, is essential for expression of a subset of late-S-phase-specific genes in Saccharomyces cerevisiae. Mol Cell Biol 19: 3312–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Park AR, Cho SK, Yun UJ, Jin MY, Lee SH, Sachetto-Martins G, Park OK (2001) Interaction of the Arabidopsis receptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. J Biol Chem 276: 26688–26693 [DOI] [PubMed] [Google Scholar]
- Pawson T (2003) Organization of cell-regulatory systems through modular-protein-interaction domains. Philos Transact Ser A Math Phys Eng Sci 361: 1251–1262 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnick DM, Halazonetis TD (2000) Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature 406: 430–435 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K, Russinova E, Gadella TW Jr, Willemse J, De Vries SC (2002) The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev 16: 1707–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavridi ES, Huyen Y, Loreto IR, Scolnick DM, Halazonetis TD, Pavletich NP, Jeffrey PD (2002) Crystal structure of the FHA domain of the Chfr mitotic checkpoint protein and its complex with tungstate. Structure 10: 891–899 [DOI] [PubMed] [Google Scholar]
- Stone JM, Collinge MA, Smith RD, Horn MA, Walker JC (1994) Interaction of a protein phosphatase with an Arabidopsis serine-threonine receptor kinase. Science 266: 793–795 [DOI] [PubMed] [Google Scholar]
- Stone JM, Trotochaud AE, Walker JC, Clark SE (1998) Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol 117: 1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Hsiao J, Fay DS, Stern DF (1998) Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281: 272–274 [DOI] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124: 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Goto N, Okada K, Takahashi H (2002) Hydrotropism in abscisic acid, wavy, and gravitropic mutants of Arabidopsis thaliana. Planta 216: 203–211 [DOI] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, Burtner C, Odden AR, Young K, Taylor NE, et al (2003) Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res 13: 524–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, et al (1998) Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93: 467–476 [DOI] [PubMed] [Google Scholar]
- Vulsteke V, Beullens M, Boudrez A, Keppens S, Van Eynde A, Rider MH, Stalmans W, Bollen M (2004) Inhibition of spliceosome assembly by the cell cycle-regulated protein kinase MELK and involvement of splicing factor NIPP1. J Biol Chem 279: 8642–8647 [DOI] [PubMed] [Google Scholar]
- Willemsen V, Wolkenfelt H, de Vrieze G, Weisbeek P, Scheres B (1998) The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125: 521–531 [DOI] [PubMed] [Google Scholar]
- Williams RW, Wilson JM, Meyerowitz EM (1997) A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc Natl Acad Sci USA 94: 10467–10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Zhu JK (2002) Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J Biol Chem 277: 8588–8596 [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Smerdon SJ (2001) PhosphoSerine/threonine binding domains: you can't pSERious? Structure 9: R33–R38 [DOI] [PubMed] [Google Scholar]
- Yongkiettrakul S, Byeon IJ, Tsai MD (2004) The ligand specificity of yeast Rad53 FHA domains at the +3 position is determined by nonconserved residues. Biochemistry 43: 3862–3869 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.