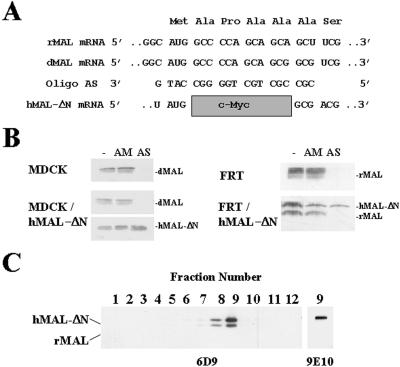
Figure 1.
Depletion of endogenous MAL by transfection with an antisense phosphorothioate oligonucleotide. (A) Nucleotide sequence of the oligonucleotides used. The sequence of the antisense oligonucleotide AS, used in MAL depletion experiments, and its alignment with the rMAL, dMAL, and hMAL-ΔN mRNA species are shown. Note that the deleted sequence in hMAL-ΔN mRNA prevents pairing with oligonucleotide AS. (B) Top panels, transfection of oligonucleotide AS in MDCK and FRT cells causes a drop in endogenous MAL protein levels. Cells were transfected with AM or AS oligonucleotide or not and incubated at 37°C. After 48 h, cell extracts were subjected to immunoblot analysis with anti-MAL mAb 2E5 (MDCK cells) or 6D9 (FRT cells). Bottom panels, hMAL-ΔN is resistant to depletion by oligonucleotide AS. Cells stably expressing hMAL-ΔN were transfected with AM or AS oligonucleotide and incubated at 37°C. After 48 h, cell extracts were subjected to immunoblot analysis with anti-MAL mAb 2E5 and anti-tag mAb 9E10 (MDCK cells) or anti-MAL 6D9 mAb (FRT cells), which recognizes both endogenous (rMAL) and exogenous (hMAL-ΔN) MAL species. (C) An intact amino terminus is not required for incorporation of hMAL into GEMs. FRT cells stably expressing a c-Myc-tagged hMAL protein bearing a deletion of the four amino acids adjacent to the initial methionine residue (hMAL-ΔN) were lysed with 1% Triton X-100 at 4°C, and the extract was centrifuged to equilibrium. After fractionation from the bottom of the tube, aliquots from each fraction were analyzed by immunoblotting with mAb 6D9. An aliquot of fraction 9 from the gradient was analyzed by immunoblotting with anti-tag mAb 9E10 to identify unambiguously tagged hMAL-ΔN. Note that because of the 6-amino-acid difference (10 amino acids from the c-Myc tag minus 4 amino acids from the deleted region) hMAL-ΔN migrates slower than rMAL.