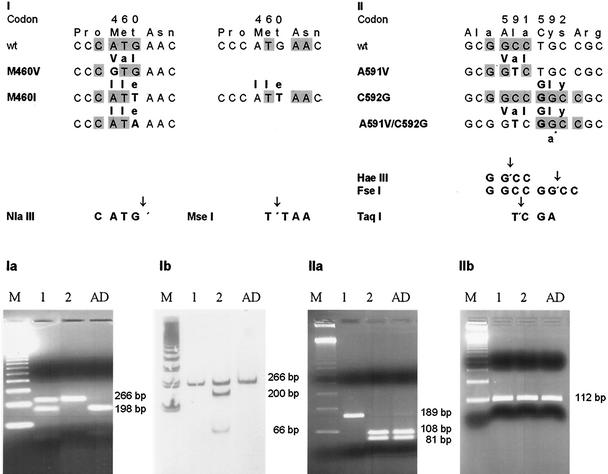
FIG. 1.
(I) Discrimination of the ATG-to-GTG/ATA versus the ATG-to-ATT mutation in UL97 codon 460. (Ia) NlaIII digest: sample 1 (tracheal secretion of a kidney transplant recipient) shows a mixture of wild-type and mutant strains (lane 1), and sample 2 (PP6, GCV-resistant laboratory strain) shows the presence of a mutant strain only (lane 2). (Ib) MseI digest with respect to NlaIII: sample 1 shows the presence of a wild-type strain (the presence of a ATG-to-GTG or -ATA mutation can be assumed) (lane 1); sample 2 shows mixture of wild-type and mutant strains (ATG to ATT must exist, and ATG to GTG or ATA can be assumed) (lane 2). Sequencing revealed a GTG and wild-type mixture for sample 1 and an ATT and GTG mixture for sample 2. (II) Pitfall of the 591/592 double mutation: use of an HaeIII digest for A591V screening can be misleading when there also exists a C592G mutation; an HaeIII digest as well as an FseI digest will result in wild-type patterns. Using the 591/592R mismatch primer (a*), a new TaqI site will be generated. (IIa) HaeIII digest: sample 1 (leukocyte, +251 days post PBSCT) shows only the mutant strain (lane 1); sample 2 (cerebrospinal fluid, + 263 days post PBSCT) shows only the wild-type strain (lane 2). (IIb) TaqI digest: exclusion of a 591V/592G double mutation. Lanes AD, AD169 GCV-sensitive laboratory strain; lanes M, 123-bp ladder.