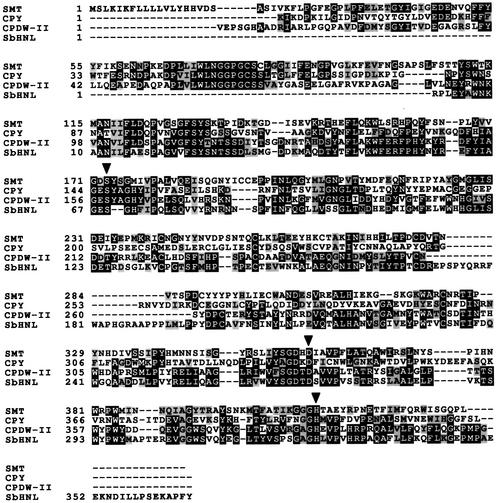
Figure 6.
Alignment of the SNG1 Gene Product with Serine Carboxypeptidases and SCPL Proteins.
An alignment of SMT with the yeast carboxypeptidase Y (CPY), wheat carboxypeptidase (CPDW-II), and the hydroxynitrile lyase from Sorghum bicolor (SbHNL) (only an incomplete sequence was available in the database) was prepared by using the ClustalW algorithm. Amino acids that are identical in two or more proteins are shaded in black, and conservative amino acid substitutions are shaded in gray. Putative active residues in SMT (S-173, D-358, and H-411) are designated with black arrowheads, based on their alignment with the carboxypeptidase Y catalytic triad. Dashes denote gaps introduced to optimize the amino acid alignment.