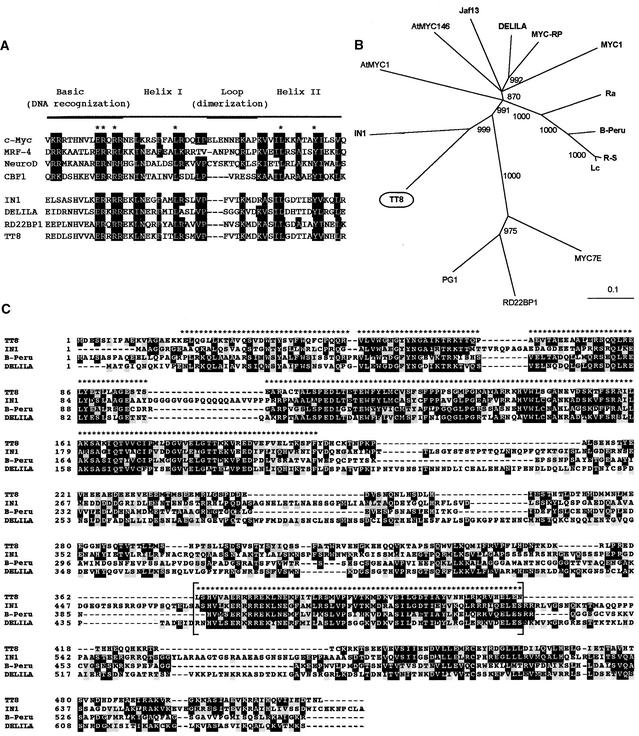
Figure 5.
Deduced TT8 Amino Acid Sequence and Comparison with bHLH-Related Protein Sequences.
(A) Amino acid sequence comparison of the bHLH regions encoded by TT8 and several MYC-related proteins. Asterisks indicate amino acid residues found in MYC-related protein sequences from all eukaryotic organisms, and conserved amino acids are boxed in black. Dashes indicate gaps inserted to improve the alignment. Sequences shown are those from human c-MYC (GenBank accession number X00198), newt MRF-4 (X82836), Xenopus NeuroD (U28067), yeast CBF1 (M33620), maize IN1 (U57899), Antirrhinum DELILA (M84913), Arabidopsis RD22BP1 (AB000875), and Arabidopsis TT8 (this study).
(B) Distance analysis of several plant bHLH-related factor sequences recovered by using a BLAST algorithm on the GenBank and EMBL databases. The following bHLH protein sequences were used to build the tree (accession numbers are in parentheses): maize Lc (M26227), maize R-S (X15806), maize B-Peru (X57276), maize IN1 (U57899), maize MYC7E (AF061107), rice Ra (U39860), pea PG1 (U18348), Gerbera MYC1 (AJ007709), bryophyte MYC-RP (AB024050), Antirrhinum DELILA (M84913), petunia Jaf13 (AF020545), and Arabidopsis AtMYC1 (D83511), Arabidopsis AtMYC146 (AF013465), Arabidopsis RD22BP1 (AB000875), and Arabidopsis TT8 (this study). Sequences were aligned using ClustalX and manually adjusted. For tree construction, only the N terminus region and the bHLH domain were used, as marked in (C). The consensus tree presented was obtained by neighbor-joining analysis, bootstrapped with 1000 iterations by ClustalX, and drawn with the TreeView program. Bootstrap values are indicated at each branchpoint; branches with a bootstrap score <850 were eliminated. Relative branch length (0.1) is indicated below the tree. Proteins in boldface have been reported to be involved in plant flavonoid metabolism.
(C) Sequence comparison between TT8 and three other bHLH proteins involved in plant flavonoid pigmentation. Identical amino acids are boxed in black, and similar amino acids are boxed in gray. Brackets delimit the bHLH region. Sequences used are from maize IN1 (GenBank accession number U57899), maize B-Peru (X57276), Antirrhinum DELILA (M84913) and Arabidopsis TT8 (this study). Asterisks above the sequences indicate amino acid residues used to build the distance tree shown in (B). Dashes were introduced to optimize alignment.