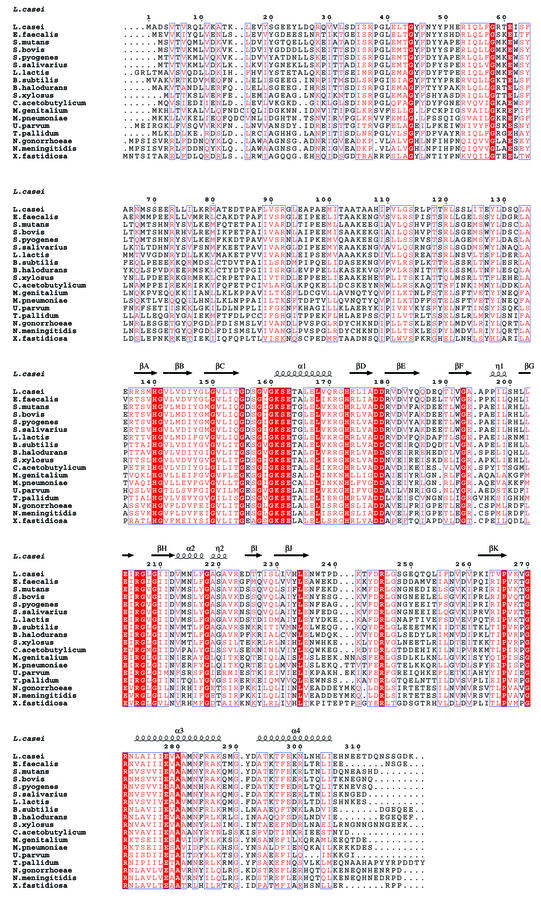
Fig. 1. Alignment of bacterial HprK/P sequences. The secondary structure on top is defined by DSSP (Kabsch and Sander, 1983) for the truncated L.casei subunit. Blue frames are for conserved residues, white characters in red boxes for strict identity, and red characters in white boxes for similarity. Accession numbers for Gram-positive bacteria: Lactobacillus casei SwissProt Q9RE09, Enterococcus faecalis SwissProt O07664, Streptococcus mutans SwissProt Q9ZA56, Streptococcus bovis SwissProt Q9WXK7, Streptococcus pyogenes orf1717 (http://pedant.mips.biochem.mpg.de), Streptococcus salivarius SwissProt Q9ZA98, Lactococcus lactis MOLOKO database (http://spock.jouy.inra.fr), Bacillus subtilis Swiss-Prot O34483, Bacillus halodurans TrEMBL P82557, Staphylococcus xylosus SwissProt Q9S1H5, Clostridium acetobutylicum (http://www.genomecorp.com/htdocs/sequences/clostridium/clospage.html), Mycoplasma genitalium SwissProt P47331, Mycoplasma pneumoniae Swiss-Prot P75548, Ureaplasma parvum SwissProt Q9PR69. Gram-negative bacteria: Treponema pallidum SwissProt O83600, Neisseria gonorrhoeae (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html), Neisseria meningitidis TrEMBL Q9K6Y4, Xylella fastidiosa SwissProt Q9PDH3. Figure realized with ESPript (Gouet et al., 1999).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.