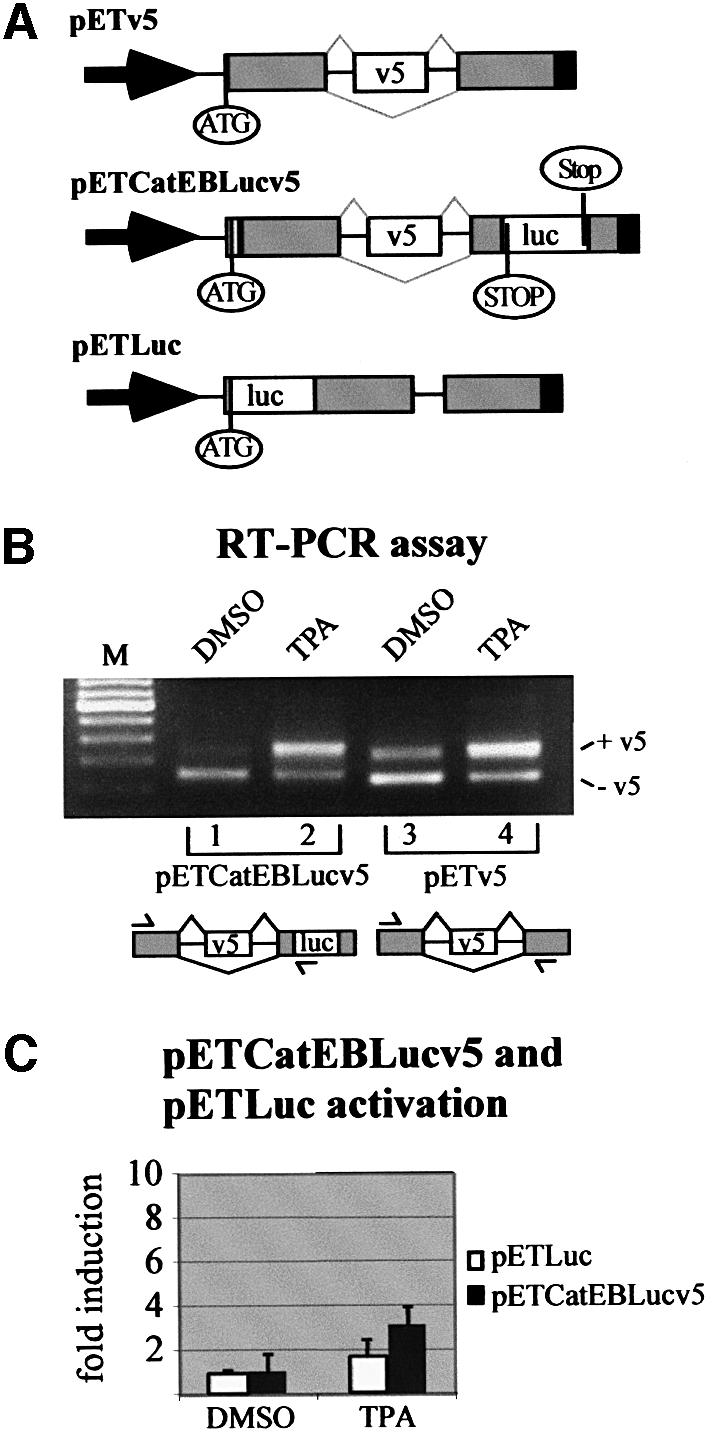
Fig. 3. A luciferase-based splice reporter assay for exon inclusion of CD44 v5. (A) Schematic representations of the original pETv5 splice reporter construct (König et al., 1998), the luciferase splice reporter construct (pETCatEBLucv5) and the construct used as a control for transcriptional activation (pETLuc). The scheme for pETCatEBLucv5 illustrates the principle of the assay. Skipping of the v5 exon sequence leads to a reading frame that ends before the luciferase coding sequence (STOP). Inclusion of the v5 exon generates a reading frame for a luciferase fusion protein, ending after the luciferase coding sequence (Stop). Arrow, RSV LTR promoter; ATG, start codon for the fusion protein; gray boxes, insulin exon 2 and 3 sequences; open boxes, CD44 exon v5, and luciferase sequence, respectively; black boxes, SV40 poly(A) signal. Thin black lines, intron sequences; gray lines indicate splice pattern. (B) RT–PCR analysis of LB-17 cells transiently transfected with splice reporter constructs. Cells were harvested and assayed 24 h after transfection with either 2 µg of the splice reporters pETCatEBLucv5 (lanes 1 and 2) or pETv5 (König et al., 1998) (lanes 3 and 4) and treatment with DMSO or TPA (40 ng/ml) for 6 h as indicated. RT–PCR analysis of minigene transcripts resulted in a short fragment (pETv5: 244bp; pETCatEBLucv5: 253 bp) when CD44 exon v5 was skipped, and in a large fragment (pETv5: 354 bp; pETCatEBLucv5: 370 bp) when CD44 exon v5 was included. Identity of PCR fragments was confirmed by sequencing. Schematic drawings of the splice reporter constructs indicating the positions of the primers (arrows) used for PCR are given below the corresponding lanes (see legend to A). M, DNA molecular size marker (100 bp ladder; Gibco-BRL). (C) Detection of luciferase activity. LB-17 cells were transiently transfected with 2 µg of the luciferase splice reporter construct pETCatEBLucv5 or the control reporter for promoter activation, pETLuc. Eighteen hours after transfection, the cells were treated with TPA (40 ng/ml) or DMSO for 6 h before they were harvested and subjected to luciferase assays. Luciferase light unit counts of stimulated cells were divided by counts measured in unstimulated cells to obtain values of fold induction. Each bar represents data of three independent experiments and standard deviations are indicated on top of the bars.
