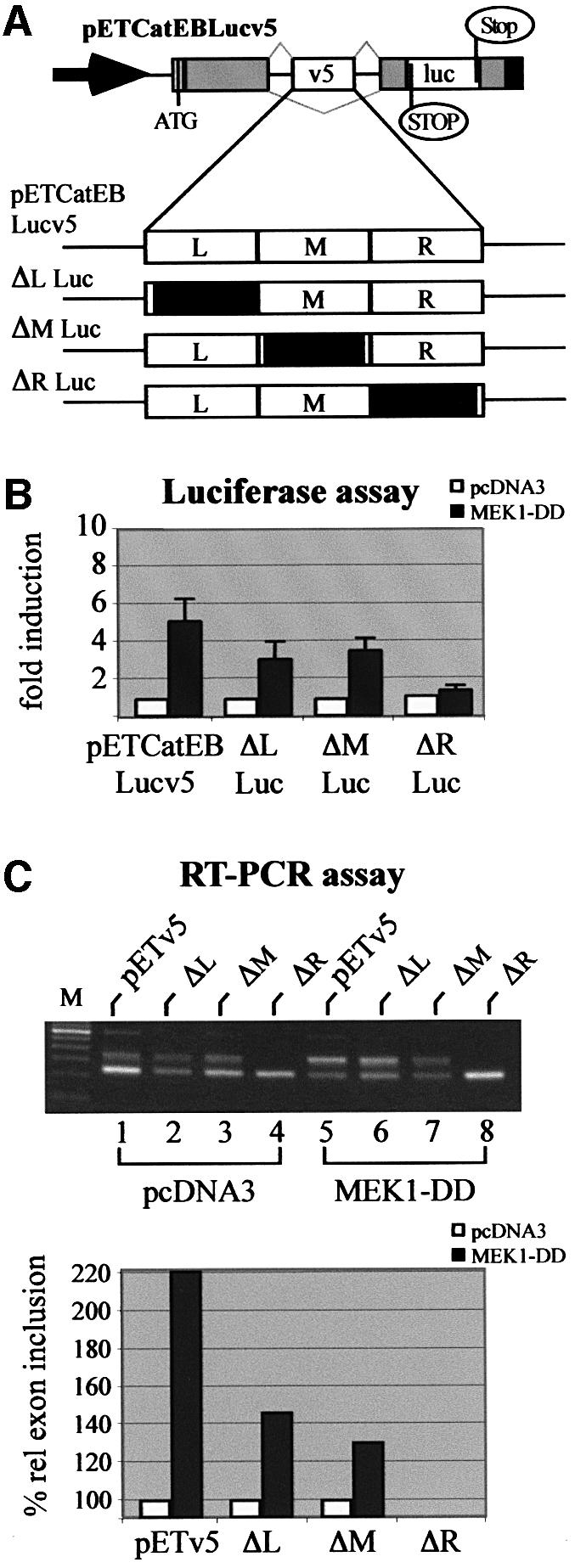
Fig. 7. Silencer sequences of CD44 v5 respond to activation of the ERK pathway. (A) Scheme of the wild-type CD44 v5 luciferase splice reporter and the mutants used in (B) and (C). Arrow, RSV LTR promoter; ATG, start codon for the fusion protein; gray boxes, insulin exon 2 and 3 sequences; open boxes, CD44 exon v5, and luciferase sequence, respectively; black boxes, SV40 poly(A) signal. Thin black lines, intron sequences; gray lines indicate splice pattern. Black boxes indicate the part (left, L; middle, M; or right, R) of the CD44 exon v5 that was replaced by a heterologous sequence (König et al., 1998). (B) MEK-induced usage of wild-type and mutant CD44 exon sequences. Two micrograms of the wild-type or the indicated mutant splice reporter were co-transfected with 2 µg of a plasmid expressing MEK1-DD (filled bars), or with the empty expression plasmid pcDNA3 (open bars) into LB-17 cells. Cells were harvested and assayed for luciferase activity 24 h after transfection. (C) RT–PCR analysis of CD44 v5 exon inclusion. LB-17 cells were co-transfected with 2 µg of wild-type and mutant splice reporters and 2 µg of constitutively active MEK or vector control (pcDNA3). Twenty-four hours after transfection cells were harvested and RT–PCR analysis was performed. Quantification of RT–PCR bands was performed as described in the legend to Figure 4B.
