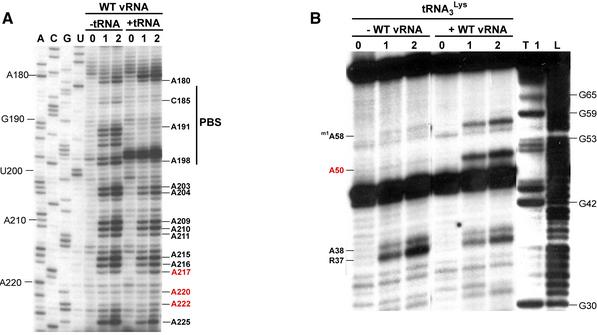
Figure 5.
Structural probing of the HIV-1 Hxb2 RNA-tRNA3Lys complex. (A) Comparison of the DMS modification profiles of nucleotides 180–225 of wild type Hxb2 vRNA, in its free form and engaged in the primer–template complex. Lanes marked 0, 1 and 2 correspond to a mock reaction without DMS and modification for 5 and 10 min, respectively. Lanes ACGU correspond to sequencing reactions. (B) Comparison of the DEPC modification profile of tRNA3Lys, either free or involved in the primer–template complex. Lanes marked 0, 1 and 2 correspond to a mock reaction without DEPC and modification for 20 and 40 min, respectively. Lanes T1 and L1 correspond to a sequencing reaction with RNase T1, which is specific for G residues, and a ladder generated by mild alkaline hydrolyis, respectively. Nucleotides whose reactivity in the primer-template complex are inconsistent with the existence of the PAS–antiPAS interaction are indicated by red labels.