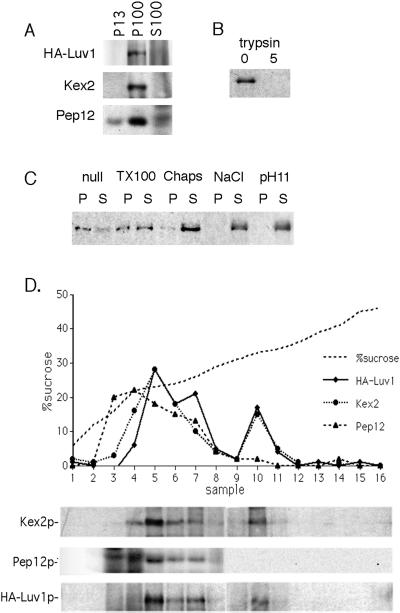
Figure 5.
Biochemical analysis of Luv1p. (A) Luv1p sediments at 100,000 × g. Protein extract was prepared from cells containing CEN-based HA-LUV1 (YMC3), and total lysates were sedimented sequentially to give a 13,000 × g pellet (P13), a 100,000 × g pellet (P100), and a supernatant (S100). Fractions were separated by SDS-PAGE and analyzed by Western blotting with the use of anti-HA, anti-Kex2p, and anti-Pep12p antibodies (see MATERIALS AND METHODS). (B) Luv1p is protease accessible. Total cell lysates from HA-Luv1p–expressing cells were treated with 0 and 5 μg of trypsin for 30 min at 25°C before analysis as in A. (C) Luv1p solubility. Total cell lysates from HA-Luv1p–expressing cells were incubated on ice for 30 min alone (null) or with 2% Triton X-100, 2% 3-([3-chloramidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate (CHAPS), 1 M NaCl, or 0.1 M Na2CO3 (pH 11) before sedimentation at 150,000 × g to give pellet (P) and supernatant (S) fractions. Fractions were analyzed as in A. (D) Luv1p cofractionates with Kex2p. HA-Luv1p–expressing cells (YMC3) were lysed, and S13 fractions were prepared and subjected to equilibrium sedimentation through a sucrose gradient. Fractions were analyzed by SDS-PAGE, followed by Western blotting with anti-HA, anti-Kex2p, and anti-Pep12p antibodies. The intensities of protein bands are shown in the graph, and the sucrose concentrations of the recovered fractions are also indicated.