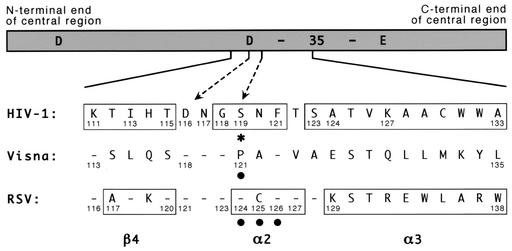
FIG. 2.
Amino acid numbering for the central region of different integrases. A schematic of the central domain of HIV-1 integrase is shown with the locations of the 3 acidic residues of the active-site D,D-35-E motif indicated (the number 35 refers to the spacing between the last two conserved residues of this motif). Amino acid sequences of the indicated region from HIV-1, visna virus, and RSV are shown, with dashes indicating identity to the HIV-1 sequence. Numbers that mark landmarks, or are referred to in the text or in this legend, are provided. Boxes indicate the β4 strand, α2 helix, and α3 helix as defined in the original papers that described the HIV-1 and RSV crystal structures (4, 9). The dashed arrows highlight HIV-1 residues D116 and S119; the latter also is marked with an asterisk. Solid circles indicate amino acids in the visna virus and RSV proteins for which substitutions were made. The HIV-1 sequence shown and used for these studies is that of BH10, which is related to HXB2 but has an S instead of G at position 123 and a K instead of R at position 127 (35, 36). It differs from the NL4-3 strain used for the HIV-1 crystal structure described by Dyda et al. (9) by having an I rather than V at position 113 and an A rather than T at position 124. The source of the visna virus and RSV integrases have been described previously (41), and the sequences presented are invariant between known strains.