Abstract
The latent membrane protein 1 (LMP-1) oncoprotein of Epstein-Barr virus (EBV) is a constitutively active, CD40-like cell surface signaling protein essential for EBV-mediated human B-cell immortalization. Like ligand-activated CD40, LMP-1 activates NF-κB and Jun kinase signaling pathways via binding, as a constitutive oligomer, to tumor necrosis factor receptor-associated factors (TRAFs). LMP-1's lipid raft association and oligomerization have been linked to its activation of cell signaling pathways. Both oligomerization and lipid raft association require the function of LMP-1's polytopic multispanning transmembrane domain, a domain that is indispensable for LMP-1's growth-regulatory signaling activities. We have begun to address the sequence requirements of the polytopic hydrophobic transmembrane domain for LMP-1's signaling and biochemical activities. Here we report that transmembrane domains 1 and 2 are sufficient for LMP-1's lipid raft association and cytostatic activity. Transmembrane domains 1 and 2 support NF-κB activation, albeit less potently than does the entire polytopic transmembrane domain. Interestingly, LMP-1's first two transmembrane domains are not sufficient for oligomerization or TRAF binding. These results suggest that lipid raft association and oligomerization are mediated by distinct and separable activities of LMP-1's polytopic transmembrane domain. Additionally, lipid raft association, mediated by transmembrane domains 1 and 2, plays a significant role in LMP-1 activation, and LMP-1 can activate NF-κB via an oligomerization/TRAF binding-independent mechanism. To our knowledge, this is the first demonstration of an activity's being linked to individual membrane-spanning domains within LMP-1's polytopic transmembrane domain.
Epstein-Barr virus (EBV) is a human tumor virus associated with a number of malignancies (26). EBV's target in vivo is the B lymphocyte. Infected primary B lymphocytes are stimulated to leave G0 and cycle indefinitely by activation of cell signaling pathways (31, 32). The activation process results from a combination of receptor-mediated signaling triggered by virus binding to CR2 on the B lymphocyte and mimicking of B-cell activation signaling pathways by viral gene products. Thus, EBV has evolved to take advantage of existing signaling pathways in B cells that culminate in B-cell activation and differentiation (33).
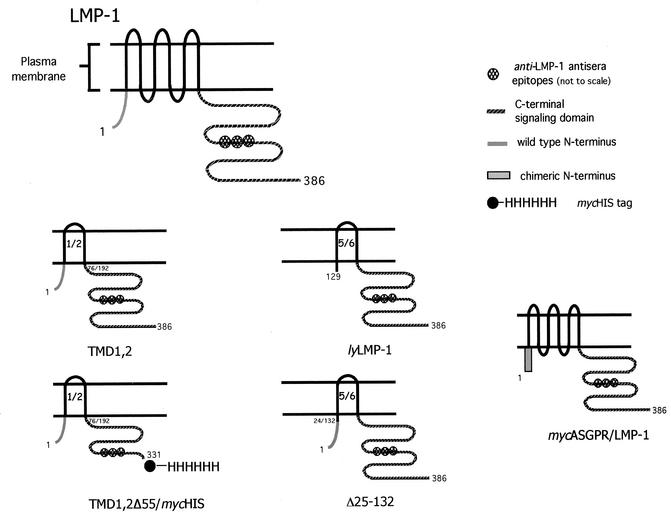
One viral “mimic” is the latent membrane protein 1 (LMP-1), a latent gene product essential for B-cell immortalization by EBV. LMP-1 is a polytopic membrane protein with six membrane-spanning segments fused to intracellular amino and carboxy termini (N and C termini) (Fig. 1). LMP-1 signal transduction closely resembles that of CD40/CD40 ligand in that LMP-1's and CD40's carboxy-terminal signaling domains bind cellular tumor necrosis factor receptor-associated factor (TRAF)/tumor necrosis factor receptor-associated death domain (TRADD) proteins and activate NF-κB and Jun kinase (JNK) pathways, resulting in cell survival and proliferation (6, 10). The TRAF/TRADD binding C termini of the two signaling proteins are in fact interchangeable (7, 16).
FIG. 1.
Schematic of LMP-1 and LMP-1 variants used in this study: comparison of full-length LMP-1, TMD1,2, TMD1,2Δ55/mycHis, lyLMP-1, Δ25-132, and mycASGPR/LMP-1. TMD1,2 lacks TMDs 3 to 6; TMD1,2Δ55/mycHis is a Myc- and His-tagged deletion of TMD1,2 lacking residues 331 to 386 of LMP-1; lyLMP-1 lacks the cytoplasmic N terminus and first four TMDs (residues 1 to 128); Δ25-132 is essentially lyLMP-1 with the wild-type LMP-1 cytoplasmic N terminus fused to TMD 5 (15); mycASGPR/LMP-1 is a chimera in which LMP-1's cytoplasmic N terminus was replaced with a Myc-tagged heterologous N terminus from the H1 asialoglycoprotein receptor (ASGPR) (4).
Importantly, unlike CD40, whose activation is ligand dependent, LMP-1 activation is constitutive. Both ligand-stimulated CD40 and LMP-1 are found in “lipid rafts,” as defined operationally by their fractionation with detergent-resistant membranes and colocalization with known lipid raft-associated signaling proteins (3, 17, 19). CD40 activation or LMP-1 expression results in TRAF3 recruitment to lipid rafts, suggesting the formation of a lipid raft-organized signaling complex (17, 19). Recent results support a model in which LMP-1's constitutive activation results either from formation of a ligand-independent, oligomerized TRAF/TRADD binding domain composed of an unknown number of LMP-1 monomers, from LMP-1's lipid raft association, or from a combination of both (19). Targeting LMP-1's C terminus to lipid rafts by tagging with the 10-amino-acid myristoylation/palmitoylation sequence from the Yes oncoprotein activates C-terminal NF-κB signaling (19). Interestingly, although TRAF3 is localized with LMP-1 in lipid rafts, TRAF2 and TRAF1, critical players in C-terminal signaling, are not detectably colocalized with LMP-1 in lipid rafts (17). It is not yet clear how lipid raft targeting regulates LMP-1's signaling activity. Both lipid raft association and oligomerization require LMP-1's N terminus/transmembrane domain (TMD) (14, 17, 19). However, whether lipid raft association is required for oligomerization or vice versa has yet to be determined. Together, these results are consistent with a model in which LMP-1 signaling complexes consist of higher-order LMP-1 oligomers to which signaling proteins bind and whose activity is regulated by lipid raft association.
Signals transduced by LMP-1 to NF-κB and JNK activation minimally require LMP-1's cytoplasmic C-terminal signaling domain so long as it is either linked to a transmembrane and extracellular domain, rendering it ligand dependent (or cross-linkable), or targeted to lipid rafts (11, 14, 19). LMP-1's N terminus-transmembrane domain (N terminus/TMD) contributes to LMP-1 signaling in a complex manner. This domain is essential for positioning the C-terminal signaling domain in the cytosol to allow coupling to intracellular signaling molecules (4). More importantly, the N terminus/TMD plays a critical and sufficient role in LMP-1 homo-oligomerization and lipid raft targeting, both of which contribute to constitutive activation of signaling (14, 17, 19). Intriguingly, replacement of CD40's extracellular domain/TMD with LMP-1's N terminus/TMD results in a more active NF-κB signaling complex than that resulting from activation of wild-type CD40 by CD40 ligand (19). Thus, LMP-1's N terminus/TMD constitutes a more potent activating domain than does CD40's ligand-activated extracellular domain/TMD. These findings suggest strongly that LMP-1's TMD must possess properties, in addition to the oligomerization/lipid raft targeting properties shared by CD40, that contribute to the efficiency of constitutive LMP-1 signaling.
LMP-1 exerts a cytostatic effect on the cell when expressed at elevated levels (12, 15, 18). This negative growth regulation results from induction of cytostasis rather than cell death (12, 18). Negative growth regulation by LMP-1 was initially recognized as inhibition of cell proliferation and detected by the failure of LMP-1 transfectants to produce progeny (15, 18). LMP-1-induced cytostasis does not require LMP-1's previously identified C-terminal signaling activities but rather depends solely on the N terminus/TMD (18). Thus, cytostasis represents an LMP-1 activity that is independent of signaling functions elicited by LMP-1's C terminus. Without a cytoplasmic N-terminal sequence, the TMD cannot activate cytostasis (4, 18). A positively charged cytoplasmic N terminus of unrelated sequence but of similar charge and size can replace LMP-1's cytoplasmic N-terminal sequence in cytostasis, indicating that the absolute sequence of the cytoplasmic N terminus is not a critical determinant (4). LMP-1's TMD has been proposed to promote cytostasis via interaction with and regulation of putative signaling molecules (i.e., receptors) in the plasma membrane in a manner akin to the E5 oncoprotein's specific association with and regulation of the platelet-derived growth factor receptor (5, 18). The relationship between LMP-1's cytostatic activity and lipid raft association/oligomerization is not understood except for the fact that all three activities require LMP-1's N terminus/TMD.
Taken together, these results strongly suggest that LMP-1's multimembrane-spanning TMD plays an active role in LMP-1 signaling, a role beyond that of a simple membrane anchor. LMP-1's TMD contributes to constitutive activation of C-terminal signaling via two key activities, oligomerization and lipid raft association. Furthermore, the TMD is a critical activator of negative growth-regulatory signaling, independent of C-terminal signaling. These findings support a model in which LMP-1's TMD contributes, via distinct and separable activities, to LMP-1's constitutive C-terminal signaling and negative growth regulation.
Because of the complex and intriguing contribution of LMP-1's TMD to all aspects of LMP-1 signaling, we have begun an in-depth analysis of the contribution of the TMD to LMP-1 function. We found that the first and second TMDs, in the absence of TMDs 3 to 6 and when preceded by the N-terminal cytoplasmic tail, are sufficient to activate cytostasis and lipid raft targeting. TMDs 1 and 2 retained the ability to activate C-terminal signaling but not as potently as LMP-1. Of particular interest is the inability of this minimal TMD to mediate homo-oligomerization and TRAF binding. Our results show that TMDs 1 and 2 and TMDs 5 and 6 are not functionally equivalent and identify TMDs 1 and 2 as a key subdomain harboring a subset of the functions of LMP-1's larger TMD. Furthermore, we provide evidence that oligomerization and lipid raft association are separable activities of LMP-1.
MATERIALS AND METHODS
Plasmids.
pCMV-TMD1,2, is a pcDNA3-based vector encoding LMP-1's cytoplasmic amino terminus, TMDs 1 and 2, and the C-terminal signaling domain (Fig. 1). Sequences following TMDs 1 and 2 and preceding the cytoplasmic C terminus were deleted by PCR. The resulting sequence encodes amino acids 1 to 76 and 192 to 386 in frame, with no heterologous residues at the junction. pCMV-LMP-1 and pCMV-lyLMP-1 are pcDNA3-based vectors encoding LMP-1 and the N-terminally truncated LMP-1 protein (referred to as lyLMP-1), respectively (4, 9). Δ25-132 is a deletion mutant of LMP-1 lacking TMDs 1 to 4 and has been described previously (15). mycASGPR/LMP-1 is an LMP-1 chimera in which the wild-type LMP-1 cytoplasmic N terminus has been replaced with a Myc-tagged N terminus from the H1 asialoglycoprotein receptor (ASGPR) (4).
pRSV-LMP-1, pRSV-TMD1,2, and pRSV-lyLMP-1 are pRc-RSV-based vectors (Invitrogen). pCMV-LMP-1myc was constructed from pCMV-LMP-1 and encodes the 10-amino-acid Myc epitope at LMP-1's C terminus. CΔ55 encodes the LMP-1 mutant CΔ55, which lacks the last 55 codons from the C terminus (24). NΔ25 encodes the LMP-1 deletion mutant NΔ25, which lacks the first 25 codons from the N terminus (25). pCMV-TMD1,2Δ55/mycHis and pRSV-TMD1,2Δ55/mycHis encode a C-terminal deletion of TMD1,2 lacking LMP-1's last 55 amino acids in Myc-His expression vectors (Invitrogen); TMD1,2Δ55/mycHis encodes both Myc and His epitopes at its C terminus. p1242 is a luciferase reporter plasmid encoding the luciferase gene driven by the minimal fos promoter with three upstream κB binding sites from the major histocompatibility complex class I gene (gift of Bill Sugden [27]). pCMV-TRAF1/HA-1, pCMV-TRAF2/HA-1, and pCMV-TRAF3/HA-1 encode hemagglutinin (HA)-1-tagged human TRAF1, TRAF2, and TRAF3 and were gifts from B. Sugden (29).
Antibodies.
Anti-LMP-1 antiserum is an affinity-purified rabbit polyclonal serum raised against LMP-1's C terminus (residues 188 to 352) fused to glutathione S-transferase (8). Monoclonal antibodies recognizing the Myc (9E10) and HA-1 (F-7) epitopes were from Santa Cruz Biochemicals. Anti-rabbit immunoglobulin secondary antibodies conjugated to horseradish peroxidase and fluorescein isothiocyanate were from Promega and Sigma, respectively.
Cells and transfections.
293 cells are an adherent human embryonic kidney carcinoma cell line grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum; HEp2 is an adherent cell line grown in Dulbecco's modified Eagle's medium with 10% calf serum. Cells were electroporated with a Bio-Rad gene pulser and harvested 1 to 2 days posttransfection (4).
Western analysis.
Transfected cells were harvested and lysed in 4× sodium dodecyl sulfate (SDS) sample buffer, boiled, and resolved on 10% acrylamide gels (4). Proteins were transferred to Immobilon (Millipore), and blocked for 1 h in phosphate-buffered saline-5% milk-0.1% Tween 20. Blots were then incubated with primary antibody (1:4,000 anti-LMP-1 antiserum) and secondary antibody (1:5,000 horseradish peroxidase-conjugated anti-rabbit immunoglobulin antibody). Blots were visualized by enhanced chemiluminescence according to the manufacturer's instructions (Amersham).
Flotation assays.
293 cells (107) were electroporated in duplicate with 5 μg of cytomegalovirus (CMV)-based LMP-1 expression vectors and harvested 2 days posttransfection. Lipid raft association was assayed as described previously (17). Samples were solubilized in MNE buffer (25 mM MES [morpholineethanesulfonic acid, pH 6.5], 150 mM NaCl, 5 mM EDTA) containing 0.2% Triton and protease inhibitors (aprotinin, pepstatin, leupeptin, and phenylmethylsulfonyl fluoride) and homogenized with 10 strokes of a Dounce homogenizer on ice. Samples were mixed 1:1 in MNE containing 80% sucrose, placed in a 5-ml ultracentrifuge tube, and overlaid with 2 ml of 30% sucrose-MNE and then with 1 ml of 5% sucrose-MNE. Samples were centrifuged at 40,800 rpm in an SW50.1 rotor (Beckman) for 18 h at 4°C. Gradients were fractionated by taking 400-μl fractions (14 total) from the top of the gradient, which were then added to 200 μl of 4× SDS sample buffer. Extracts were resolved by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to Immobilon, and stained with anti-LMP-1 antiserum.
Cytostasis assay.
Cytostasis assays were performed as described previously (4) with a modification of the procedure reported by Kaykas and Sugden (18). HEp2 cells were transfected by electroporation and immediately plated at clonal density in six-well plates on coverslips. Cells were grown for 96 h and fixed in acetone-methanol (1:1) for 20 min at −20°C. Coverslips were blocked in phosphate-buffered saline-1% calf serum and stained with 1:50 anti-LMP-1 antiserum followed by 1:40 fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin secondary antibody (Sigma). Coverslips were mounted on slides with 4′,6′-diamidino-2-phenylindole (DAPI) mounting medium. Slides were visualized under a Nikon Eclipse E800 fluorescent microscope (40× objective), and images were captured with a Cooke SensiCam digital camera with SlideBook software. All colonies on a given coverslip were scored for the number of positive (LMP-1-immunoreactive) cells and expressed as number of positive cells per colony. A total of 500 colonies were scored for each transfection.
Activation of NF-κB.
Subconfluent 293 cells were cotransfected by electroporation in duplicate with 2.0 μg and 7.0 μg of RSV-LMP-1 expression vectors and 1.0 μg of p1242 (luciferase gene under the control of three κB binding sites upstream of a minimal fos promoter [27]). Cells were harvested 48 h posttransfection, and extracts were assayed for luciferase activity with the Dual Light assay kit from Tropix. Luciferase values were averaged for each sample, and data are expressed as a percentage of maximal LMP-1 activation. Data shown are representative of four independent experiments.
Oligomerization assay.
293 cells transfected with CMV-based vectors were harvested 48 h posttransfection, and lysates were prepared for immunoprecipitation as described by Coffin et al. (4). Anti-Myc (9E10) antibody was used as the immunoprecipitating antibody. Immunoprecipitates were recovered with protein G-agarose (Boehringer), and beads were washed four times in 1× radioimmunoprecipitation assay buffer. The final pellet was resuspended and boiled in 4× SDS sample buffer. Immunoprecipitates were resolved by SDS-PAGE and transferred to Immobilon. Blots were stained with anti-LMP-1 antiserum and visualized by enhanced chemiluminescence as described above.
TRAF binding assay.
293 cells were electroporated with CMV-based expression vectors and harvested and lysed as described for the oligomerization assay except that 1× complete protease inhibitor (Boehringer Mannheim) was added to the TNA lysis buffer (50 mM Tris [pH 7.4], 0.15 mM NaCl, 1% NP-40, 5 mM EDTA). Anti-HA-1 antibody F-7 was used as the immunoprecipitating antibody. Immunoprecipitates were recovered and analyzed by Western blot as described above.
RESULTS
Generation of TMD1,2.
To begin to dissect the contribution of individual TM domains to LMP-1 function, we generated an LMP-1 deletion mutant lacking TMDs 3 to 6 (Fig. 1). TMD1,2 encodes LMP-1's cytoplasmic N terminus and TMDs 1 and 2 fused to the C-terminal signaling domain. TMD1,2 is similar in overall structure to the naturally occurring LMP-1 variant (lyLMP-1) in that both proteins encode a pair of TMDs fused to LMP-1's cytoplasmic C terminus. However, TMD1,2 encodes the cytoplasmic N terminus and the first pair of TMDs (1 and 2), whereas lyLMP-1 encodes the last pair of TMDs (5 and 6). Unlike TMD1,2, lyLMP-1 initiates at codon 129 of the LMP-1 open reading frame and thus is lacking the cytoplasmic N terminus. Proteolytic analysis of lyLMP-1 in intact cells demonstrated that, like LMP-1, lyLMP-1's C terminus was inside the cell (not shown). Additional controls used to identify activities associated with TMDs 1 and 2 included Δ25-132, which is essentially identical to lyLMP-1 but encodes the wild-type LMP-1 cytoplasmic N terminus fused to TMD 5 (15), and mycASGPR/LMP-1, in which LMP-1's cytoplasmic N terminus is replaced with a Myc-tagged N terminus from the H1 asialoglycoprotein receptor (ASGPR) (4) (Fig. 1).
TMD1,2 was assayed for wild-type transmembrane insertion by chymotrypsin cleavage in intact cells (4). Chymotrypsin cleaves LMP-1 in the first extracellular loop between TMDs 1 and 2 and has been used previously to demonstrate that the first loop is exposed to the outside of the cell (4, 21, 23). TMD1,2 was oriented across the plasma membrane, as is LMP-1 (N and C termini within the cell, hydrophilic loop between TMDs 1 and 2 outside the cell) (not shown). We next used TMD1,2 to determine if TMDs 1 and 2 contribute to or are sufficient for LMP-1's signaling activities and biochemical properties.
TMD1,2 associates with lipid rafts.
LMP-1 signaling activity is correlated with its localization in the plasma membrane, specifically in lipid raft microdomains, where LMP-1 is thought to form an active signaling complex (17, 19). LMP-1's cytoplasmic N terminus/TMD is required for lipid raft association (17), but the sequences within LMP-1's TMD contributing to lipid raft association have not been identified. We therefore asked whether TMDs 1 and 2 were sufficient for lipid raft targeting. lyLMP-1, which does not home to lipid rafts (17), and Δ25-132, which is identical to lyLMP-1 but with LMP-1 residues 1 to 24 (cytoplasmic N terminus) fused to TMD 5, were used as controls for TMD1,2 in this assay. lyLMP-1, Δ25-132, and TMD1,2 are similarly hydrophobic (i.e., all three possess two TMDs) and have the same large C terminus (Fig. 1).
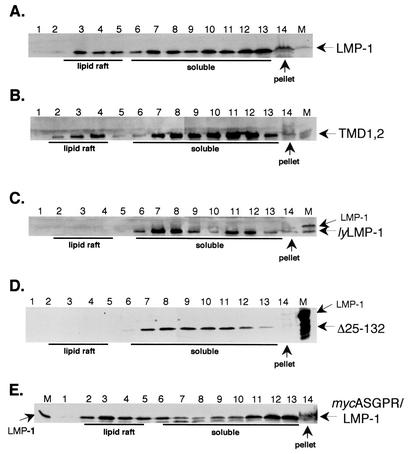
Human 293 cells were transfected with wild-type LMP-1, TMD1,2, lyLMP-1, Δ25-132, and mycASGPR/LMP-1 expression vectors and assayed 2 days posttransfection for lipid raft association with a sucrose flotation assay (17). Triton lysates were subjected to flotation through discontinuous sucrose gradients, the gradients were fractionated, and the fractions were assayed by Western blot (Fig. 2). Lipid rafts floated toward the top of the gradient (fractions 2 to 5), Triton-soluble material remained in the lower layer of the gradient (fractions 6 to 13), and Triton-insoluble material was pelleted (fraction 14). Consistent with the results of Higuchi et al. (17), LMP-1 was found in the lipid raft fractions (fractions 3 to 4) as well as in the Triton-soluble fractions (fractions 7 to 13) and the Triton-insoluble fraction (pellet). The sucrose flotation profiles of TMD1,2 and LMP-1 were indistinguishable in that both proteins were clearly present in the lipid raft fraction. In contrast, lyLMP-1 was excluded from lipid rafts, as reported previously (17). Importantly, Δ25-132 was indistinguishable from lyLMP-1 and mycASGPR/LMP-1 was indistinguishable from wild-type LMP-1 in their sucrose flotation profiles, ruling out a role for LMP-1's cytoplasmic N terminus in lipid raft homing. TMD1,2, lyLMP-1, and Δ25-132 were detected at low levels or not at all in the Triton-insoluble pellet, whereas LMP-1 and mycASGPR/LMP-1 were easily detectable. These results demonstrate that TMDs 1 and 2 of LMP-1 but not the cytoplasmic N terminus or TMDs 5 and 6 possess lipid raft targeting activity.
FIG. 2.
Lipid raft association of TMD1,2. 293 cells were transfected with pCMV-based LMP-1, TMD1,2, lyLMP-1, Δ25-132, and mycASGPR/LMP-1 expression vectors. Two days posttransfection, cells were solubilized in a buffer containing 0.2% Triton X-100, homogenized, mixed with an equal volume of 80% sucrose, and overlaid with 30% and 5% sucrose in MNE buffer. Following centrifugation for 18 h at 200,000 × g, fractions were taken from the top of each gradient and resolved on SDS-PAGE gels. Gels were transferred to Immobilon and stained with anti-LMP-1 antiserum. A marker for each LMP-1 protein is shown on the right or left of each blot (transfected 293 cell whole-cell lysate, lane M), and arrows point to the indicated LMP-1 protein. (A) LMP-1, (B) TMD1,2, (C) lyLMP-1, (D) Δ25-132, (E) mycASGPR/LMP-1. The lipid raft, soluble, and pellet fractions are labeled beneath. These results are representative of three independent experiments.
Cytostasis induction by TMD1,2.
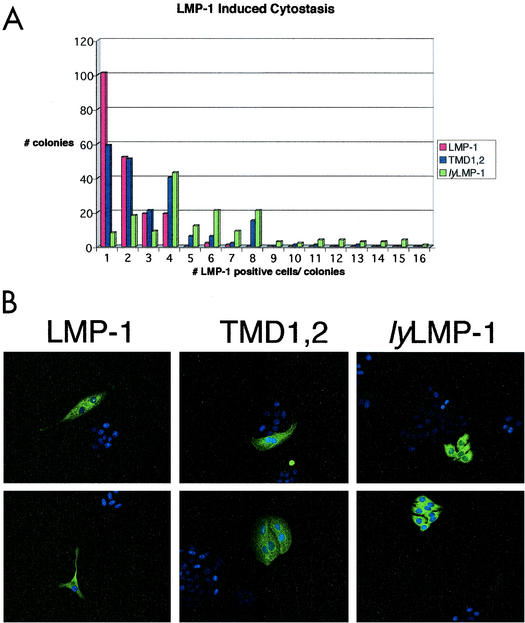
LMP-1 has long been known to negatively regulate cell growth when expressed at elevated levels in cells. We next asked if TMDs 1 and 2 play a role in LMP-1-mediated cytostasis by using a modification of the clonal assay described by Kaykas and Sugden (18). HEp2 cells were transfected with expression vectors encoding LMP-1, TMD1,2, and lyLMP-1. Immediately following transfection, cells were plated at clonal density and allowed to grow. Four days after plating, cells were fixed and stained with anti-LMP-1 antiserum and DAPI. Colonies were scored for the number of LMP-1-immunoreactive cells (Fig. 3).
FIG. 3.
TMD1,2-induced cytostasis. HEp2 cells were transfected with pCMV-LMP-1, pCMV-TMD1,2, or pCMV-lyLMP-1 and plated on coverslips at clonal density immediately following transfection. Four days postplating, cells were fixed and stained with anti-LMP-1 antiserum. (A) All colonies on a given coverslip were scored for the number of positive cells (LMP-1 immunoreactive) and data are expressed as number of positive cells per colony. Data shown as “1 LMP-1-positive cell per colony” include both individual immunoreactive cells (see part B, LMP-1) and single immunoreactive cells within a colony (see upper panel in part B, TMD1,2). A total of 500 colonies were scored for each transfection. (B) Immunofluorescent images of representative colonies are shown for each transfection; two representative images are shown for each of the introduced plasmids; the name of the introduced plasmid is shown above each equivalent set of images (upper and lower panels). These results are representative of three separate experiments. Both LMP-1- and TMD1,2-positive HEp2 colonies primarily have one to four LMP-1-immunoreactive cells, whereas lyLMP-1-positive colonies primarily have four to eight immunoreactive cells. LMP-1- and TMD1,2-positive cells tend to be multinucleated and much larger than lyLMP-1-positive cells.
A histogram showing the number of immunoreactive cells per colony versus colony number is shown in Fig. 3A. The majority of LMP-1- and TMD1,2-expressing colonies consisted of one to four immunoreactive cells, and most of these cells were much larger than their nonimmunoreactive neighbors or lyLMP-1-expressing cells and were frequently multinucleated. The majority of lyLMP-1-expressing colonies consisted of four to eight immunoreactive cells. Importantly, LMP-1- and TMD1,2-expressing colonies with greater than eight immunoreactive cells per colony were never seen, whereas colonies with 9 to 16 lyLMP-1-expressing cells per colony were detected. lyLMP-1-expressing cells were much smaller than LMP-1- and TMD1,2-expressing cells and resembled untransfected neighboring cells. Overall, TMD1,2 behaved more like the cytostatic LMP-1 than the noncytostatic lyLMP-1 in this assay. If the total number of colonies for each colony size category are added, 50% of all LMP-1-expressing colonies and 34% of all TMD1,2-expressing colonies had one immunoreactive cell, compared to 4% of all lyLMP-1-expressing colonies. Furthermore, if immunoreactive single-cell colonies are distinguished from multicell colonies with one immunoreactive cell (the numbers in the histogram do not distinguish these categories), then ≈31% of all LMP-1-positive colonies consisted of a single immunoreactive cell, compared to ≈23% for TMD1,2 and ≈2% for lyLMP-1 (not shown).
Colonies with four immunoreactive cells were observed most frequently for lyLMP-1. In addition, the morphology of LMP-1- and TMD1,2-immunoreactive cells was indistinguishable (very large cells, frequently multinucleated) and distinct from that of lyLMP-1. Western analysis showed that the levels of expressed LMP-1, TMD1,2, and lyLMP-1 were equivalent (not shown), suggesting that although TMD1,2 was cytostatic, it was less efficient than LMP-1 in this regard. It is important to note that Δ25-132, which is equivalent to lyLMP-1 except that it encodes the N-terminal cytoplasmic domain fused to TMD 5 (see Fig. 1), is not cytostatic (15). Furthermore, mycASGPR/LMP-1, which encodes a heterologous cytoplasmic N terminus, exhibits nearly wild-type cytostatic activity (4). These results demonstrate that TMDs 1 and 2 alone can replace LMP-1's hydrophobic TMD in induction of cytostasis and strongly suggest that the cytoplasmic N terminus of LMP-1 does not contribute to this activity except to position the TMDs properly in the membrane.
NF-κB activation by TMD1,2.
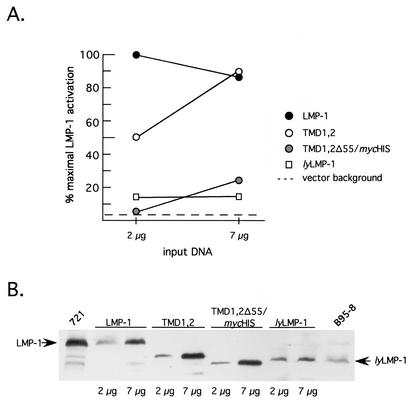
LMP-1, like CD40, activates NF-κB via binding cellular TRAF proteins to its C terminus (6). Since NF-κB activation by LMP-1 requires the TMD, we next asked if TMDs 1 and 2 were sufficient for NF-κB activation. Cells were transfected with LMP-1 expression vectors and an NF-κB responsive luciferase reporter (4). TMD1,2 activated NF-κB, but its concentration dependence differed from that of LMP-1. At low levels of expression (comparable to LMP-1 levels expressed in the 721 lymphoblastoid cell line), TMD1,2 resembled the inactive variant lyLMP-1 in its activity (Fig. 4 and data not shown (4, 9, 27). At higher levels of expression, TMD1,2 activated NF-κB to the same high level as did LMP-1. LMP-1 activation but not TMD1,2 activation was biphasic under the conditions of our assay. NF-κB activation by LMP-1, as monitored by reporter assays, has been shown previously to be biphasic with respect to the LMP-1 expression level: lower LMP-1 levels stimulate and higher LMP-1 levels inhibit NF-κB activation (30).
FIG. 4.
Activation of NF-κB by TMD1,2 in 293 cells. 293 cells were cotransfected with RSV-LMP-1 expression vectors encoding LMP-1, lyLMP-1, TMD1,2, or TMD1,2Δ55/mycHis (2 μg and 7 μg), together with an NF-κB-responsive luciferase reporter as described previously (4) and in Materials and Methods. (A) Extracts were assayed for luciferase activity 48 h posttransfection with a Dual Light assay kit (Tropix). Data are expressed as a percentage of maximal LMP-1 activation, with maximal activation by LMP-1 occurring at 2 μg of input DNA (423,310 relative light units [RLU]). The activation detected in the presence of 7 μg of empty vector control (pRC-RSV) is denoted by the dotted line. Solid circles, LMP-1; open circles, TMD1,2; shaded circles, TMD1,2Δ55/mycHis; open squares, lyLMP-1. (B) Extracts from A were assayed for LMP-1 expression by Western blot. Each lane was loaded with 5 × 104 cell equivalents. The expressed protein is shown above the blot, and lanes are marked below the blot with input DNA amounts (2 μg or 7 μg). The amount of LMP-1 protein detected in each lane is representative of duplicate transfections in a single experiment. Arrows point to LMP-1 and lyLMP-1. 721 cells (5 × 105 cells) and tetradecanoyl phorbol acetate- and butyrate-induced B95-8 cells (105 cells) (EBV-positive lymphoblastoid cell lines) were loaded on either side of the blot and served as markers for LMP-1 and lyLMP-1, respectively. Shown is a representative of three independent experiments. TMD1,2 activates NF-κB with a different concentration dependence than LMP-1 via a mechanism involving LMP-1 residues 331 to 386, which include CTAR2.
Δ25-132, like lyLMP-1, is inactive in this assay (27), whereas mycASGPR/LMP-1 is indistinguishable from wild-type LMP-1 (4). Thus, TMDs 1 and 2 alone conferred constitutive activation upon LMP-1's C terminus but with a potency that differed from that of the complete TMD. The lack of activity of TMD1,2Δ55/mycHis (described in Fig. 1) in this assay indicated that activation by TMD1,2 was mediated by amino acids 331 to 386 of LMP, the C-terminal region encoding CTAR2.
Oligomerization of TMD1,2 with itself and with LMP-1.
LMP-1's TMD is essential for homo-oligomerization. We next asked whether the first two TMDs were sufficient to mediate oligomerization by assessing the ability of TMD1,2 to oligomerize with LMP-1 by coimmunoprecipitation analysis (Fig. 5A and B). 293 cells were transfected singly (Fig. 5A) or in combination (Fig. 5B) with expression vectors encoding LMP-1myc (Myc tag at C terminus), a C-terminal deletion mutant of LMP-1 (CΔ55), an N-terminal deletion mutant of LMP-1 (NΔ25), and TMD1,2. CΔ55 oligomerizes with LMP-1, and its faster mobility in SDS gels prompted its use as a resolvable LMP-1-interacting partner (4). NΔ25 lacks the cytoplasmic N terminus but encodes the TMD and C-terminal signaling domain and does not interact with LMP-1 (4). NΔ25 therefore served as a noninteracting but hydrophobic negative control. Extracts were immunoprecipitated with anti-Myc antibody, and precipitating proteins were visualized by Western blot with anti-LMP-1 antiserum.
FIG. 5.
Oligomerization of TMD1,2. 293 cells were electroporated with the indicated CMV-based expression vectors and harvested 48 h posttransfection, and cell lysates were immunoprecipitated with an anti-Myc antibody (9E10, Santa Cruz). Immunoprecipitates (IP) and whole-cell lysates (WCL) from transfected cells were resolved on SDS-10% PAGE gels and visualized by Western blot with anti-LMP-1 antiserum as described previously (4) and in Materials and Methods. The introduced LMP-1 protein is shown above each blot, and arrows on the sides of the blots mark each protein's migration. (A) Immunoprecipitation of individually expressed proteins with anti-Myc antibody. (B) Coimmunoprecipitation of LMP-1myc with CΔ55 (LMP-1 deletion mutant lacking 55 amino acids from the C terminus, included as an LMP-1-interacting partner [positive control]), NΔ25 (LMP-1 deletion mutant lacking the cytoplasmic N terminus, included as a noninteracting control), or TMD1,2. (C) Coimmunoprecipitation of TMD1,2 with TMD1, 2Δ55mycHis (a C-terminally truncated TMD1,2 lacking 55 amino acids from the C terminus in pcDNA3.1/mycHis [Invitrogen]). TMD1,2 does not detectably interact with LMP-1 or with itself.
The only individually expressed protein immunoprecipitated with anti-Myc antibodies was LMP-1myc (Fig. 5A). As predicted, CΔ55 coprecipitated with coexpressed LMPmyc, whereas NΔ25 did not (Fig. 5B). TMD1,2 was undetectable in LMP-1myc/TMD1,2 coprecipitates, demonstrating that TMD1,2 did not oligomerize with LMP-1. To ask whether TMD1,2 could interact with itself, a C-terminally truncated and Myc-His epitope-tagged mutant of TMD1,2, TMD1,2Δ55/mycHis (lacking residues 331 to 386, as in CΔ55), was employed (Fig. 1). The two TMD1,2 mutants were expressed singly or together and immunoprecipitated with Myc antibodies (Fig. 5C). TMD1,2Δ55/mycHis was easily resolved from TMD1,2 on SDS gels and efficiently precipitated with Myc antibodies. However, TMD1,2 was not coprecipitated from extracts of cells coexpressing TMD1,2Δ55/mycHis and TMD1,2. These results, together with the results in Fig. 5A and B, demonstrate that TMD1,2 does not interact detectably with full-length LMP-1 or with itself and suggest strongly that TMDs 1 and 2 do not mediate oligomerization in the absence of TMDs 3 to 6. This was unexpected, given the fact that TMD1,2 retained the ability to activate NF-κB (Fig. 4).
TRAF binding of TMD1,2.
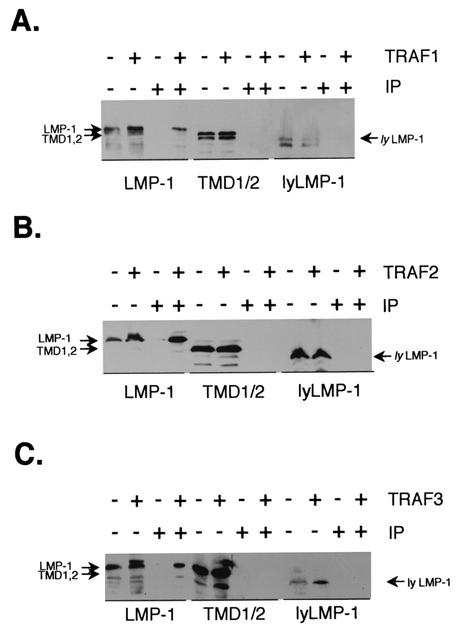
Oligomerized TRAFs bind to trimerized tumor necrosis factor receptor superfamily members (25, 28), and by analogy, LMP-1 is thought to bind to TRAFs as a constitutive oligomer. Therefore, TMD1,2's inability to oligomerize with itself or LMP-1 suggested that it would be unable to bind to TRAF proteins. To test whether TMD1,2 could interact with TRAF proteins, 293 cells were transfected with the LMP-1, TMD1,2, or lyLMP-1 expression vector together with either TRAF1, TRAF2, or TRAF3 (all tagged with the HA-1 epitope from influenza virus hemagglutinin). Extracts were immunoprecipitated with anti-HA antibodies, and coprecipitating LMP-1 proteins were visualized by Western blot with anti-LMP-1 antiserum (Fig. 6A to C). As expected, LMP-1 was detectable in TRAF1, TRAF2, and TRAF3 immunoprecipitates. Consistent with previous results, lyLMP-1 was not present in TRAF3 immunoprecipitates (9) and similarly was not detectable in TRAF1 or TRAF2 immunoprecipitates. Significantly, TMD1,2 was undetectable in TRAF1, TRAF2, and TRAF3 immunoprecipitates despite its ample expression in cotransfected cells (Fig. 6). These results demonstrate, perhaps not surprisingly, given that TMD1,2 does not oligomerize, that TMD1,2 does not interact detectably with TRAF1, TRAF2, or TRAF3.
FIG. 6.
Interaction of TMD1,2 with TRAF1, TRAF2, and TRAF3. 293 cells were transfected with CMV-based expression vectors encodingLMP-1 or TMD1,2, with or without pCMV-TRAF1/HA-1 (A), pCMV-TRAF2/HA-1 (B), or pCMV-TRAF3/HA-1 (C). Cells were harvested 24 h posttransfection, and extracts were immunoprecipitated with anti-HA-1 antibodies (F-7; Santa Cruz). TRAF immunoprecipitates (IP) and whole-cell lysates of transfected cells (WCL) were resolved by SDS-10% PAGE and analyzed by Western blot with anti-LMP-1 antiserum. The introduced LMP-1 proteins are shown below each blot, and inclusion of the indicated TRAF in the transfection is noted by a + above each blot (top row). Lanes containing HA-1 immunoprecipitates are noted by + above each blot, and whole-cell lysates are noted by − (bottom row). Individual LMP-1 proteins (LMP-1, TMD1,2, and lyLMP-1) are identified by arrows to the sides of each blot. LMP-1-immunoreactive bands migrating faster than LMP-1 are the result of degradation. LMP-1 and not TMD1,2 or lyLMP-1 is detected in TRAF1, TRAF2, and TRAF3 immunoprecipitates.
DISCUSSION
An LMP-1 deletion mutant (TMD1,2), encoding LMP-1's N terminus and TMDs 1 and 2 fused to the C-terminal signaling domain (Fig. 1), localizes to lipid rafts, is cytostatic, and activates NF-κB with diminished potency relative to LMP-1 but does not oligomerize with LMP-1 or bind TRAF1, TRAF2, or TRAF3. TMD1,2 is oriented across the plasma membrane, as is LMP-1 (N and C termini within the cell, loop between TMDs 1 and 2 outside; not shown). TMD1,2 is indistinguishable from wild-type LMP-1 with respect to lipid raft association (Fig. 2) and exhibits nearly wild-type cytostatic activity (Fig. 3). The sucrose flotation profiles (Fig. 2) and cytostatic activities (Fig. 3) of Δ25-132, lyLMP-1, and MycASGPR/LMP-1 demonstrate that it is LMP-1's first two TMDs, independent of the cytoplasmic N terminus, that encode a lipid raft targeting signal and the sequence information required for negative growth regulation.
LMP-1's association with lipid rafts has been shown to require the hydrophobic TMD (17, 19). LMP-1 colocalizes with TRAF3 in lipid rafts, and lipid raft association can activate C-terminal signaling (19). Interestingly, TRAF1 and -2 do not colocalize with LMP-1 in lipid rafts but are found with LMP-1 in the Triton-insoluble fractions (cytoskeletal fraction) (17). Thus, whether lipid raft-associated LMP-1 represents an active signaling complex remains to be determined. Our results with TMD1,2 strengthen the correlation between LMP-1's lipid raft association and activation of signaling. TMD1,2 does not oligomerize with itself (Fig. 5), nor does it bind TRAF1, TRAF2, or TRAF3 (Fig. 6). Surprisingly, TMD1,2 retained the ability to activate NF-κB, although with a different concentration dependence than LMP-1 (Fig. 4). Furthermore, the ability of TMD1,2 to activate NF-κB required residues 331 to 386, which encode CTAR2. These results suggest that NF-κB activation by TMD1,2 may be TRADD mediated. Experiments to detect LMP-1 interactions with TRADD were inconclusive, and thus we were unable to test binding of TMD1,2 to TRADD directly. Together, these results suggest that TMD1,2's localization to lipid rafts may be sufficient for activation of signaling.
Intriguingly, the behavior of TMD1,2 is very similar to that of the LMP-1 mutant MYPALMP-1 reported by Kaykas and Sugden (19). MYPALMP-1 encodes LMP-1's C terminus with the 10-amino-acid lipid raft targeting sequences from the Yes oncoprotein fused to its N terminus. Like TMD1,2, MYPALMP-1 localizes to lipid rafts and retains some but not all of LMP-1's NF-κB stimulating activity, does not oligomerize and, although not reported by Kaykas and Sugden, presumably does not bind TRAFs. Thus, TMDs 1 and 2 and the myristoylation/palmitoylation sequences from Yes similarly activate signaling from LMP-1's C terminus, presumably via lipid raft targeting.
Our results with lyLMP-1 and Δ25-132 (Fig. 2) demonstrate that TMDs 5 and 6, with or without the cytoplasmic N terminus, are not sufficient for lipid raft targeting. The lack of a role for the cytoplasmic N terminus in lipid raft targeting is strengthened by our results with mycASGPR/LMP-1 (Fig. 1), which is indistinguishable in function (4) and in lipid raft association (Fig. 2) from LMP-1. These results, together with our findings of wild-type localization of TMD1,2 to lipid rafts, indicate that the first two TMDs of LMP-1 are sufficient for lipid raft targeting. These results, and those reported in Fig. 3, demonstrate that individual TMDs 1 to 6 encode distinct activities involved in lipid raft association and cytostasis. TMDs 1 and 2 could encode LMP-1's lipid raft targeting sequence, or lipid raft targeting may result from lipid partitioning and could require a “pair” of LMP-1's first four TMDs. However, the inability of lyLMP-1 and Δ25-132 to localize to lipid rafts argues against the latter scenario. Further work is needed to identify the nature of the lipid raft targeting activity encoded in TMDs 1 and 2.
LMP-1 expression in a number of cell backgrounds results in inhibition of cell growth. This LMP-1-induced cell proliferation phenotype was first observed as the failure of LMP-1-transfected cells to produce progeny (15). LMP-1's negative proliferative effect has more recently been shown to result from cytostasis rather than cell death (12, 18). The level of LMP-1 in the cell that is sufficient to induce cytostasis is just twofold greater than that expressed in EBV-positive lymphoblastoid cells (18). The ability of LMP-1 to induce cytostasis is dependent on the TMD and independent of the cytoplasmic C terminus (18). Our previous results (4), and those reported here, demonstrate that TMDs 5 and 6, whether expressed with (15) or without the cytoplasmic N terminus (Fig. 3), cannot replace LMP-1's hydrophobic TMD in cytostasis induction. In contrast, TMDs 1 and 2 can replace TMDs 1 to 6 in cytostasis induction, albeit with somewhat reduced efficiency (Fig. 3). Again, differences in activity between lyLMP-1 and Δ25-132 versus TMD1,2 illustrate for the first time that the six TMDs are not functionally equivalent.
TMD1,2 activation of NF-κB differs from activation by LMP-1 (Fig. 4). Maximal activation of NF-κB by TMD1,2 is indistinguishable from that by LMP-1 in our assay, but LMP-1 activates NF-κB more potently than does TMD1,2. At low levels of expression, TMD1,2 is clearly less active than LMP-1 in NF-κB activation (Fig. 4) and resembles the inactive lyLMP-1 in its activity (not shown) (4, 9, 27). Previous results have shown that the Δ25-132 mutant is indistinguishable from lyLMP-1 in this assay (27), whereas mycASGPR/LMP-1 exhibits wild-type LMP-1 activity (4). At higher but equivalent levels of expression, LMP-1 and TMD1,2 activate NF-κB to the same level. LMP-1 but not TMD1,2, activation of NF-κB is biphasic (Fig. 4). The inhibition of gene expression observed in response to higher levels of LMP-1 requires the N terminus and TMD and may be related to LMP-1's cytostatic activity (30) or may be mediated by an as yet unidentified activity originating from LMP-1's multispanning TMD. It is possible that NF-κB activation by TMD1,2 is not biphasic because of differences between LMP-1 and TMD1,2 in efficiency of cytostasis induction (Fig. 3) or due to the fact that TMD1,2 lacks segments of LMP-1's TMD that are responsible for triggering inhibition of reporter gene activity.
Interestingly, TMD1,2 does not oligomerize with full-length LMP-1 or itself, nor does it bind TRAF1, TRAF2, or TRAF3 detectably (Fig. 5 and 6). Thus, NF-κB activation by TMD1,2 appears to be TRAF1-, TRAF2-, and TRAF3-independent. TRAF-independent NF-κB activation has also been observed for CD40 and is mediated by an unknown mechanism (1, 22). TRAF-independent NF-κB activation by CD40 is slower (i.e., takes longer to reach maximal activation) and of lower magnitude than TRAF-dependent NF-κB activation (22). The requirement for LMP-1 residues 331 to 386 in TMD1,2 activation of NF-κB suggests the possibility that CTAR2 is involved, possibly via TRADD binding. Consistent with our results is previous work showing that LMP-1's C terminus can activate NF-κB in the absence of oligomerization (and presumably TRAF binding) by addition of a lipid raft targeting signal from Yes (19). The possibility that the nonoligomerized C terminus in TMD1,2 retains low but undetectable levels of TRAF binding which are sufficient to activate NF-κB must be considered as well.
LMP-1's constitutive activation depends in part on its ability to interact with itself as an oligomer (6, 7, 11, 14, 16). Tumor necrosis factor receptor superfamily members bind oligomerized TRAF proteins via their trimerized intracellular C termini (25, 28). Likewise, LMP-1's oligomerized C terminus serves as a constitutive TRAF binding site. LMP-1 oligomerization requires the cytoplasmic N terminus/TMD, with the N terminus serving to orient the TMD correctly in the membrane and thereby promoting LMP-1/LMP-1 interactions (4, 14). Examination of the sequence of LMP-1's TMD reveals a leucine zipper-like motif in TMD 1 (LSSSLGLALLLLLLALLFWLY, residues 21 to 41 of the B95-8 sequence [2]), which could conceivably mediate interactions between LMP-1 monomers within the membrane. However, our results indicate that the first two TMDs of LMP-1, when expressed as a “pair” in the absence of TMDs 3 to 6, are not sufficient to mediate LMP-1's oligomerization. Our results with TMD1,2 are consistent with those recently reported by Kaykas et al. (20), who showed that substitution of seven leucine residues in TMD 1 with alanines had no effect on homo-oligomerization (although it impaired NF-κB activation). Furthermore, previous results argue strongly that TMDs 5 and 6 do not play a critical role in oligomerization when expressed in the absence of the first four TMDs (Δ25-132) (9, 13). It is possible that the interaction between LMP-1 monomers requires a binding interface that is only generated in the context of all six TMDs or that TMDs 3 and 4 may form the interacting site. The inability of TMD1,2 to bind TRAF1, TRAF2, or TRAF3 (Fig. 6) is not surprising given the fact that this deletion mutant does not oligomerize with itself or with LMP-1 and thus cannot form an oligomerized C-terminal TRAF binding site.
A previously unanswered question is whether LMP-1 oligomerization is required for lipid raft association or cytostasis induction as it is for constitutive activation of TRAF-dependent C-terminal signaling activities. Our results with TMD1,2, which homes to lipid rafts but does not oligomerize with LMP-1 or with itself, suggest that oligomerization is not required for lipid raft association. These findings support a model in which oligomerization and lipid raft association are distinct and separable activities of LMP-1's TMD.
These data support our working hypothesis that the TMD encodes distinct and separable activities contributing to constitutive TRAF-dependent C-terminal signaling and negative growth regulation and suggest that lipid raft targeting information is encoded in TMD 1 and/or 2. Furthermore, the difference in activity between TMD1,2 versus lyLMP-1 and Δ25-132, all of which encode two TMDs and the C terminus (Fig. 1), is strong evidence that LMP-1's six TMDs are not functionally equivalent. Our results demonstrating nonequivalence of LMP-1's TMDs are consistent with those recently reported by Kaykas et al. (20), in which TMDs 1 and 6 were shown to be functionally distinct. This is the first demonstration of an LMP-1 activity (lipid raft association and cytostasis) associated with TMDs isolated from LMP-1's polytopic TMD.
Acknowledgments
We thank Edwin Schiff for assistance in the sucrose flotation assay.
This work was supported in part by NIH grants CA-64610 and AI-01537 to J. M. Martin.
REFERENCES
- 1.Ahonen, C. L., E. M. Manning, L. D. Erickson, B. P. O. O'Conner, E. F. Lind, S. S. Pullen, M. R. Kehry, and R. J. Noelle. 2002. The CD40-TRAF6 axis controls affinity maturation and the generation of long-lived plasma cells. Nat. Immunol. 3:451-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 3.Clausse, B., K. Fizazi, V. Walczak, C. Tetaud, J. Wiels, T. Tursz, and P. Busson. 1997. High concentration of the EBV latent membrane protein 1 in glycosphingolipid-rich complexes from both epithelial and lymphoid cells. Virology 228:285-293. [DOI] [PubMed] [Google Scholar]
- 4.Coffin, W. F., III, K. D. Erickson, M. Hoedt-Miller, and J. M. Martin. 2001. The cytoplasmic amino-terminus of the latent membrane protein-1 of Epstein-Barr virus: relationship between transmembrane orientation and effector functions of the carboxy-terminus. Oncogene 20:5313-5330. [DOI] [PubMed] [Google Scholar]
- 5.DiMaio, D., C. Lai, and D. Mattoon. 2000. The platelet-derived growth factor B receptor as a target of the bovine papillomavirus E5 protein. Cytokine Growth Factor Rev. 11:283-293. [DOI] [PubMed] [Google Scholar]
- 6.Eliopoulos, A. G., and A. B. Rickinson. 1998. Epstein-Barr virus: LMP1 masquerades as an active receptor. Curr. Biol. 8:R196-R198. [DOI] [PubMed] [Google Scholar]
- 7.Eliopoulos, A. G., and L. S. Young. 1998. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene 16:1713-1742. [DOI] [PubMed] [Google Scholar]
- 8.Erickson, K. D., and J. M. Martin. 1997. Early detection of the lytic LMP-1 protein in EBV-infected B cells suggests its presence in the virion. Virology 234:1-13. [DOI] [PubMed] [Google Scholar]
- 9.Erickson, K. D., and J. M. Martin. 2000. The late lytic LMP-1 protein of Epstein-Barr virus can negatively regulate LMP-1 signaling. J. Virol. 74:1057-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrell, P. J. 1998. Signal transduction from the Epstein-Barr virus LMP-1 transforming protein. Trends Microbiol. 6:175-178. [DOI] [PubMed] [Google Scholar]
- 11.Floettmann, J. E., and M. Rowe. 1997. Epstein-Barr virus latent membrane protein-1 (LMP1) C-terminus activation region 2 (CTAR2) maps to the far C-terminus and requires oligomerisation for NF-κB activation. Oncogene 15:1851-1858. [DOI] [PubMed] [Google Scholar]
- 12.Floettmann, J. E., K. Ward, A. B. Rickinson, and M. Rowe. 1996. Cytostatic effect of Epstein-Barr virus latent membrane protein-1 analyzed with tetracycline-regulated expression in B cell lines. Virology 223:29-40. [DOI] [PubMed] [Google Scholar]
- 13.Gires, O., F. Kohlhuber, E. Kilger, M. Baumann, A. Kieser, C. Kaiser, R. Zeidler, B. Scheffer, M. Ueffing, and W. Hammerschmidt. 1999. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 18:3064-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gires, O., U. Zimber-Strobl, R. Gonnella, M. Ueffing, G. Marschall, R. Zeidler, D. Pich, and W. Hammerschmidt. 1997. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 16:6131-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerschmidt, W., B. Sugden, and V. R. Baichwal. 1989. The transforming domain alone of the latent membrane protein of Epstein-Barr virus is toxic to cells when expressed at high levels. J. Virol. 63:2469-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatzivassiliou, E., W. E. Miller, N. Raab-Traub, E. Kieff, and G. Mosialos. 1998. A fusion of the EBV latent membrane protein-1 (LMP1) transmembrane domains to the CD40 cytoplasmic domain is similar to LMP1 in constitutive activation of epidermal growth factor receptor expression, nuclear factor-kappa B, and stress-activated protein kinase. J. Immunol. 160:1116-1121. [PubMed] [Google Scholar]
- 17.Higuchi, M., K. M. Izumi, and E. Kieff. 2001. Epstein-Barr virus latent-infection membrane proteins are palmitoylated and raft-associated: Protein-1 binds to the cytoskeleton through TNF receptor cytoplasmic factors. Proc. Natl. Acad. Sci. USA 98:4675-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaykas, A., and B. Sugden. 2000. The amino-terminus and membrane-spanning domains of LMP-1 inhibit cell proliferation. Oncogene 19:1400-1410. [DOI] [PubMed] [Google Scholar]
- 19.Kaykas, A., K. Worringer, and B. Sugden. 2001. CD40 and LMP-1 both signal from lipid rafts but LMP-1 assembles a distinct, more efficient signaling complex. EMBO J. 20:2641-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaykas, A., K. Worringer, and B. Sugden. 2002. LMP-1's transmembrane domains encode multiple functions required for LMP-1's efficient signaling. J. Virol. 76:11551-11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liebowitz, D., D. Wang, and E. Kieff. 1986. Orientation and patching of the latent infection membrane protein encoded by Epstein-Barr virus. J. Virol. 58:233-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manning, E. M., S. S. Pullen, D. J. Souza, M. R. Kehry, and R. J. Noelle. 2002. Cellular responses to murine CD40 in a mouse B cell line may be TRAF dependent of independent. Eur. J. Immunol. 32:39-49. [DOI] [PubMed] [Google Scholar]
- 23.Martin, J. M., and B. Sugden. 1991. The LMP onco-protein resembles activated receptors in its properties of turnover. Cell Growth Differ. 2:653-660. [PubMed] [Google Scholar]
- 24.Martin, J. M., and B. Sugden. 1991. Transformation by the LMP oncoprotein correlates with its rapid turnover, membrane localization, and cytoskeletal association. J. Virol. 65:3246-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McWhirter, S. M., S. S. Pullen, J. M. Holton, J. J. Crute, M. R. Kehry, and T. Alber. 1999. Crystallographic analysis of CD40 recognition and signaling by human TRAF2. Proc. Natl. Acad. Sci. USA 96:8408-8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, G. (ed.). 1985. Epstein-Barr virus, first ed. Raven Press, New York, N.Y.
- 27.Mitchell, T., and B. Sugden. 1995. Stimulation of NFkB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J. Virol. 69:2968-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, Y. C., V. Burkitt, A. R. Villa, L. Tong, and H. Wu. 1999. Structural basis for self-association and receptor recognition of human TRAF2. Nature 398:533-538. [DOI] [PubMed] [Google Scholar]
- 29.Sandberg, M., W. Hammerschmidt, and B. Sugden. 1997. Characterization of LMP-1's association with TRAF1, TRAF2, and TRAF3. J. Virol. 71:4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandberg, M. L., A. Kaykas, and B. Sugden. 2000. Latent membrane protein 1 of Epstein-Barr virus inhibits as well as stimulates gene expression. J. Virol. 74:9755-9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugden, B. 1989. An intricate route to immortality. Cell 57:5-7. [DOI] [PubMed] [Google Scholar]
- 32.Sugden, B. 1994. Latent infection of B lymphocytes by Epstein-Barr virus. Semin. Virol. 5:197-205. [Google Scholar]
- 33.Thorley-Lawson, D. A., and G. J. Babcock. 1999. A model for persistent infection with Epstein-Barr virus: the stealth virus of human B cells. Life Sci. 65:1433-1453. [DOI] [PubMed] [Google Scholar]