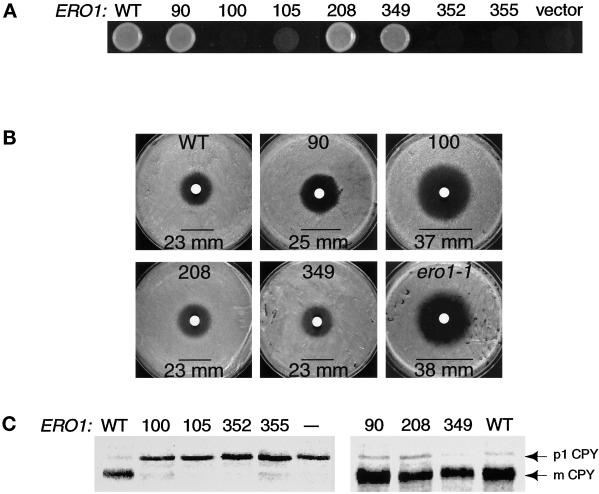
Figure 2.
Cys100, Cys105, Cys352, and Cys355 of Ero1p are important for cell viability and for oxidative protein folding in the ER. (A) Complementation of the temperature-sensitive ero1-1 mutant by alanine substitution mutants of ERO1-myc. Cells were plated on rich medium and incubated overnight at restrictive temperature (38°C). Alleles of ERO1-myc with detectable rescuing activity were expressed in an ire1Δ genetic background to prevent induction of ERO1 by the UPR. Transformants of CKY605 (ero1-1 ire1Δ::URA3) carried the following plasmids: wild type (WT), pAF85 [CEN ERO1-myc]; 90, pAF153 [CEN ero1-A90-myc]; 100, pAF154 [CEN ero1-A100-myc]; 208, pAF155 [CEN ero1-A208-myc]; or 349, pAF156 [CEN ero1-A349-myc]. Transformants of CKY559 (ero1-1) hosted the following: 105, pAF122 [CEN ero1-A105-myc]; 352, pAF96 [CEN ero1-A352-myc]; 355, pAF95 [CEN ero1-A355-myc]; or vector, pRS316 [CEN URA3]. (B) Assays for the DTT sensitivity of cells with a chromosomal deletion of ERO1 rescued by an alanine substitution mutant of ERO1-myc. Three lawns of each strain were plated on rich medium and 10 μl of 3 M DTT applied to each lawn in a filter disk. The average diameter of the zone of growth inhibition (mm) was measured after incubation at 30°C for 1.5 days. The strains shown are as follows: WT, CKY600 (ero1-Δ1-500::LEU2 p[ERO1-myc]); 90, CKY601 (ero1-Δ1-500::LEU2 p[ero1-A90-myc]); 100, CKY602 (ero1-Δ1-500::LEU2 p[ero1-A100-myc]); 208, CKY603 (ero1-Δ1-500::LEU2 p[ero1-A208-myc]); 349, CKY604 (ero1-Δ1-500::LEU2 p[ero1-A349-myc]); and ero1-1, CKY559. (C) Processing of newly synthesized CPY in ero1-1 cells expressing each alanine substitution mutant of ERO1. Cells were pulse-labeled with [35S]methionine and cysteine at 38°C for 20 min. CPY was immunoprecipitated and the samples resolved by SDS-PAGE. The ER (p1) and vacuolar (mature) forms of CPY are indicated. Samples were prepared from the same strains as in A, except that “— ” refers to untransformed CKY559.