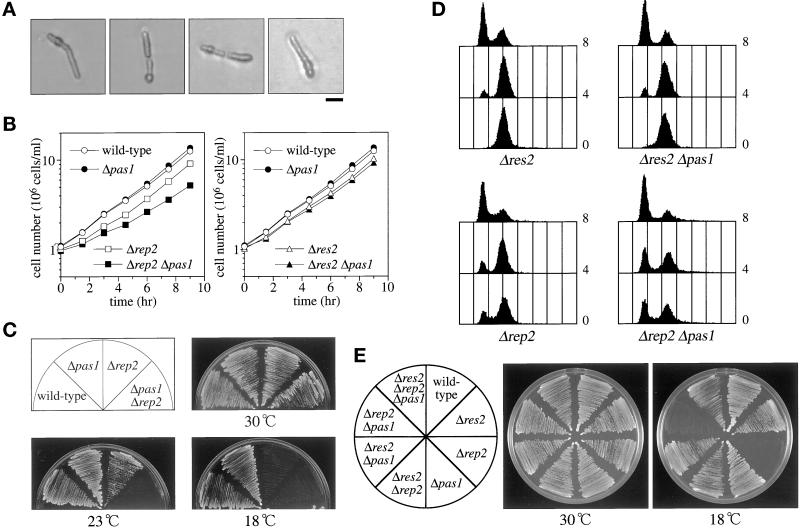
Figure 4.
Genetic interactions of pas1+ with res1+, res2+, and rep2+. (A) Terminal phenotype of Δres1 Δpas1 cells. Δres1/res1+ Δpas1/pas1+ diploid cells were tetrad dissected on YEA plates and incubated at 30°C. Cells that germinated from Δres1 Δpas1 double deletion spores (judged by genotypes of the other segregants) were photographed. Bar, 10 μm. (B) Deletion of pas1+ significantly reduces the growth rate of the Δrep2 mutant but not of the Δres2 mutant. The wild-type (L972), Δpas1 (K193-A1), Δrep2 (K166-A5), Δpas1 Δrep2 (K190-22B), Δres2 (K165-A12), and Δpas1 Δres2 (K189-3A) cells were cultured in MM (+N/2%G) at 30°C, and their growth was monitored by counting cell numbers. (C) Deletion of pas1+ greatly enhances the cold-sensitivity of the Δrep2 mutant. Cells were inoculated on MMA plates and incubated at the indicated temperatures. (D) Deletion of pas1+ enhances the slow G1 progression of the Δrep2 mutant but not that of the Δres2 mutant. Cells were grown to midlog phase at 30°C in MM (+N/2%G) and shifted to 18°C. Cells were sampled at 4 and 8 h after the temperature shift and analyzed by flow cytometry. (E) Deletion of the res2+ gene completely suppressed the cold-sensitivity of the Δpas1 Δrep2 double mutant. The wild-type (EV3A), Δres2 (M222), Δrep2 (N3-141S), Δpas1 (K182-A7), Δres2 Δrep2 (NP2-461), Δres2 Δpas1 (K189-23C), Δrep2 Δpas1 (K190-5D), and Δres2 Δrep2 Δpas1 (K539-1A) cells were inoculated on YEA plates and incubated at the indicated temperatures.