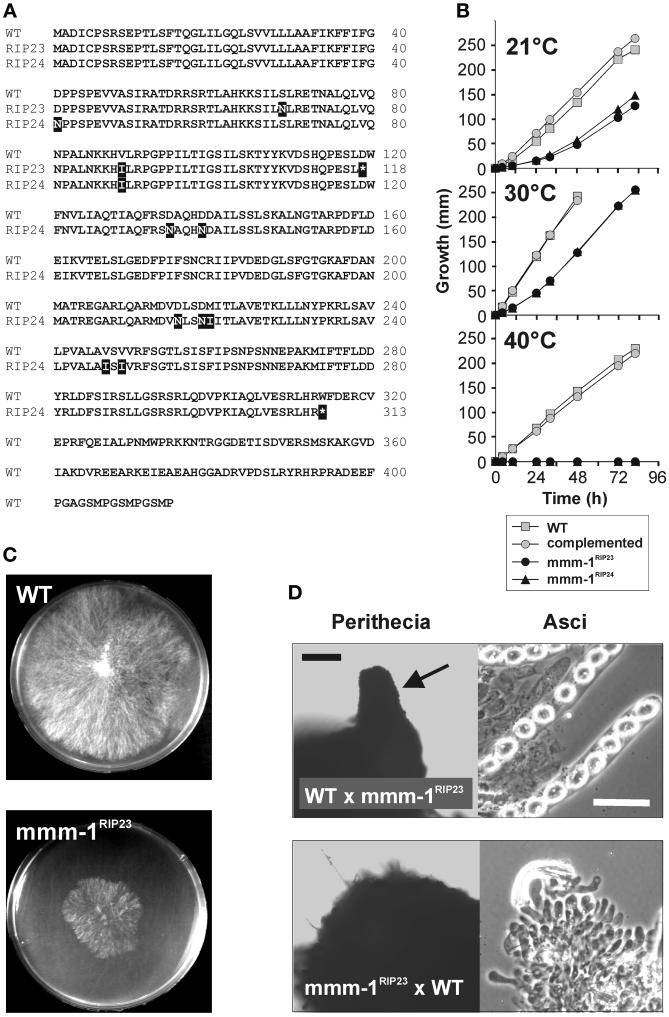
Figure 2.
Characterization of mmm-1RIP mutants. (A) Mutations in the mmm-1RIP alleles. The predicted amino acid sequences of the mmm-1 wild-type (WT) and mutant (RIP23 and RIP24) alleles were aligned. Exchanged amino acids of the mutant proteins are in black boxes, introduced translation stop codons are denoted by asterisks in black boxes. (B) Slow growth phenotype of mmm-1RIP mutants. Glass tubes containing solid growth medium were inoculated with mycelia from the wild-type strain (gray boxes), the mmm-1RIP23 mutant (black circles), the mmm-1RIP24 mutant (black triangles), and the mmm-1RIP23 mutant complemented with the wild-type mmm-1 gene (gray circles). The tubes were incubated at the indicated temperatures and the distance the mycelia had progressed along the growth medium was measured each day. One representative experiment of three is shown. (C) Colony morphology of the mmm-1RIP23 mutant. Mycelia of WT and mmm-1RIP23 were inoculated in the middle of 8-cm Petri dishes containing Vogel's medium and plates were incubated for 24 h at 37°C. (D) Female sterility of mmm-1RIP mutant. WT protoperithecia were fertilized with conidia of the mmm-1RIP23 mutant (WT × mmm-1RIP23) and mmm-1RIP23 mutant protoperithecia were fertilized with WT conidia (mmm-1RIP23 × WT) and perithecial development was allowed to occur for 14 days at 25°C. Perithecia were prepared from the mycelia and subjected to light microscopy (left). Asci were prepared by opening the perithecia with a needle (right). The beak-like structure of a mature WT perithecium is highlighted by a black arrow. Black bar, 50 μm; white bar, 20 μm.