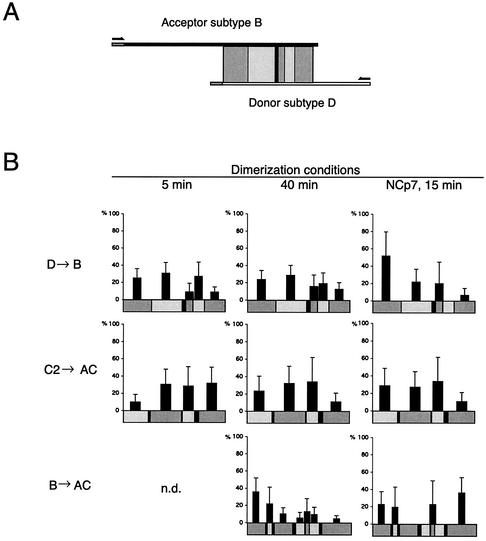
FIG. 5.
Distribution of template-switching events for different subtype combinations and conditions. (A) Illustration of the experimental setup with indication of regions of identity (shaded boxes) and phylogenetic markers (black lines) between subtypes B and D. The transcripts, primers, regions of identity, and markers are drawn to scale. (B) Distribution of template-switching events. Prior to RT transcription, the RNA substrates were dimerized for either 5 or 40 min or incubated for 15 min in the presence of NCp7. Analysis of recombinant clones was performed, and the identified template-switching events are summarized in Table 1. The y axis indicates the frequency of template switching normalized to the number of nucleotides within each region of identity. Standard error bars are estimated as Δpi = pi[(ni)−1/2 + (N)−1/2], where N is the total number of recombinants cloned and sequenced in each independent experiment and ni is the number of recombinants occurring in the interval i. All regions of identity less than 5 nucleotides long are omitted from the figure but are shown in Table 1. Three independent experiments were performed for the donor D-to-acceptor B combination (5 and 40 min) with similar distribution. All three experiments were included in the data analysis. Only single template-switching events were included. n.d., not determined.