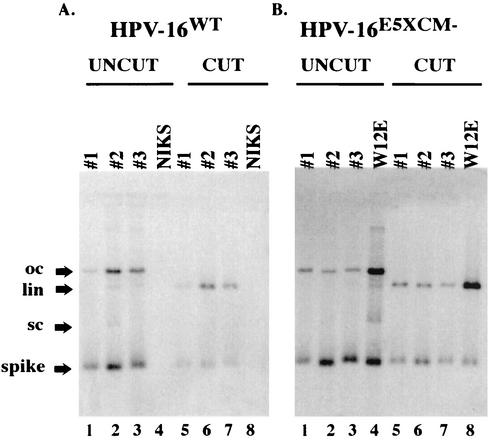
FIG. 2.
Southern analysis of low-molecular-weight (Hirt) DNA extracted from three populations each of stably transfected NIKS containing the HPV16WT (A) or HPV16E5XCM− (B) viral DNA. Hirt DNA from passage 2 populations of NIKS cells harboring HPV16WT or HPV16E5XCM− genomes (lanes 1 to 3 and 5 to 7), untransfected NIKS cells and W12E cells grown on feeders were left uncut (lanes 1 to 4) or were cut with BamHI (to linearize viral genomes) (lanes 5 to 8) and subjected to Southern analysis using an HPV16-specific probe. Arrows indicate the migration of open circular (oc), linear (lin), and supercoiled (sc) HPV16 genomes as well as a bacterially synthesized spike plasmid DNA that was added to the cells at the time of lysis to assess efficiency and reproducibility of recovery of low molecular weight DNAs using the Hirt extraction protocol.