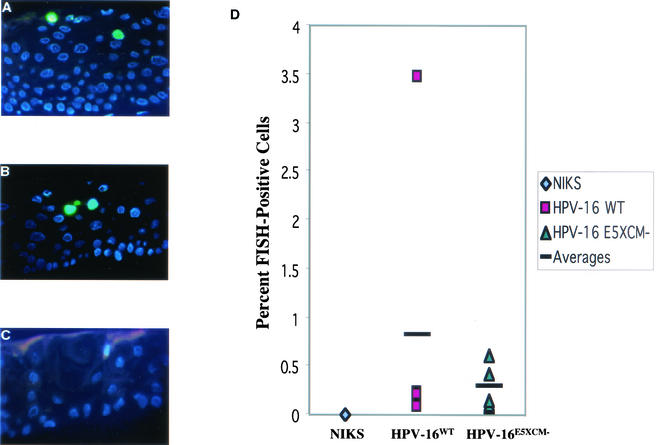
FIG. 6.
FISH analysis on sections of organotypic raft cultures of NIKS cells harboring HPV 16 WT (A) and E5 mutant (B) genomes as well as untransfected (C) NIKS cells, using an HPV16-specific probe. The sections were counterstained with DAPI (blue nuclei). FISH-positive nuclei (green), indicating amplification of viral DNA, were found in the terminally differentiating compartment of rafts generated with NIKS cells harboring either HPV16WT (A) or HPV16E5XCM− genomes (B), but not in rafts generated with untransfected NIKS (C). (D) Histogram in which the frequency of cells harboring amplified copies of HPV16 were quantified in raft cultures subjected to HPV16-specific FISH analysis, as described in Materials and Methods. No significant difference in the frequency of cells with amplified copies of HPV16 genomes was evident between the cell populations harboring WT or E5 mutant viral genomes (P = 0.8551).