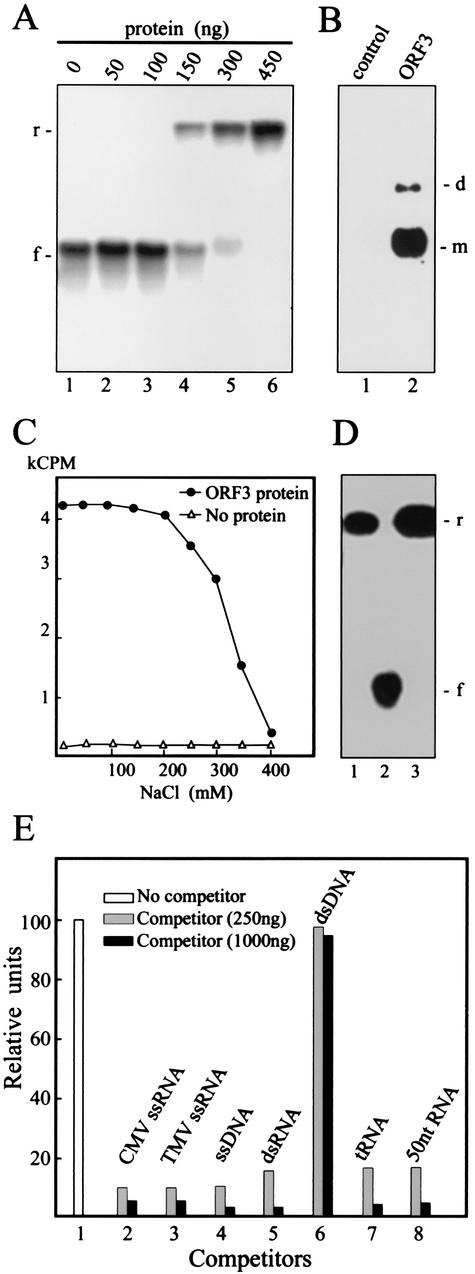
FIG. 6.
Analysis of nucleic acid binding by the ORF3-His protein. (A) Gel retardation electrophoresis assay for RNA binding by the ORF3 protein. Increasing amounts of the ORF3 protein were incubated in 15 μl of binding buffer A (see Materials and Methods) with 2ng of 32P-labeled GRV RNA transcript, and the mixtures were electrophoresed in 1% nondenaturing agarose gel. The amount of the ORF3 protein used in the assay is indicated above the lanes. The positions of free (f) and retarded (r) RNAs are shown to the left of the gel. (B) North-Western blot analysis of RNA binding by the ORF3 protein. After SDS-PAGE, the ORF3 protein was transferred onto a nitrocellulose membrane and renatured. The membrane was treated with 32P-labeled GRV RNA transcript. The ORF3 protein was substituted by BSA mixed with an equivalent protein preparation from plants infected with the empty TMV(30B) vector as a control. The positions of dimers (d) and monomers (m) are indicated to the right of the blot. (C) Salt stability of RNA-ORF3 protein complex. The ORF3 protein (450 ng) was incubated with labeled GRV RNA (4 ng) in the presence of the indicated concentrations of NaCl. After incubation, the mixtures were analyzed by nitrocellulose membrane filter-binding assay. RNA binding was quantified by determining the radioactivity of the membrane by liquid scintillation counting. kCPM, 1,000 cpm. (D) Gel retardation electrophoresis assay for RNA binding by the ORF3 protein (400 ng) as in panel A but in the presence of 350 mM (lane 1) or 200 mM (lane 3) NaCl. Lane 2 contained RNA without the ORF3 protein, preincubated in 350 mM NaCl. The positions of free (f) and retarded (r) RNAs are shown to the right of the gel. (E) Competition binding assay of the ORF3 protein to GRV ssRNA. The protein (450 ng in 15 μl) was incubated with labeled GRV RNA (4 ng) either in the absence (column 1) or in the presence of the following unlabeled competitors: CMV ssRNA (column 2), TMV ssRNA (column 3), bacteriophage M13 ssDNA (column 4), simian rotavirus SA11 dsRNA (column 5), plasmid dsDNA fragment (SmaI-digested pUC18 DNA) (column 6), tRNA (column 7), and rRNA hydrolyzed by NaOH (about 50 nucleotides in length) (column 8). The amounts of the competitors are indicated. After incubation, the mixtures were analyzed by the nitrocellulose membrane filter-binding assay. RNA binding was quantified by densitometry of autoradiographic images in relative units (100 units corresponds to RNA binding by the ORF3 protein without competitor).