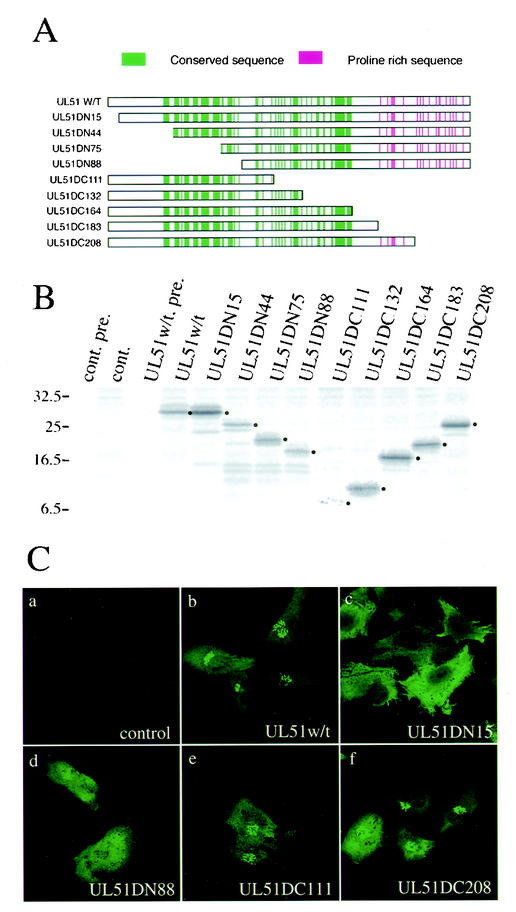
FIG. 6.
Effect of N- and C-terminal deletions on the Golgi targeting of UL51 protein. (A) Construction of the UL51 deletion mutant proteins. All of the constructs were inserted into pcDNA3.1(+) as described in Materials and Methods. Highly conserved amino acid sequences in alphaherpesviruses are shown by vertical lines in green. Proline-rich region are shown in red. (B) Expression of the UL51 deletion mutant proteins. U251 cells were transfected with pcDNA-UL51w/t or each plasmid and labeled with [35S]methionine as described in Materials and Methods. Cell lysates were subjected to immunoprecipitation with anti-UL51 rabbit polyclonal antibody, and the labeled proteins were separated by SDS-PAGE and detected by autoradiography with the Fujix Bioimaging Analyser BAS 2000 system. The main bands of each deletion protein are shown by black dots. Molecular mass markers (in kilodaltons) are shown on the left. (C) Intracellular localization of the UL51 deletion mutant proteins. U251 cells were transfected with each plasmid. After 24 h, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. The samples were stained with the anti-UL51 rabbit polyclonal antibody and then reacted with anti-rabbit IgG-conjugated FITC. Fluorescence images were obtained with the Bio-Rad MRC 1024 confocal imaging system.