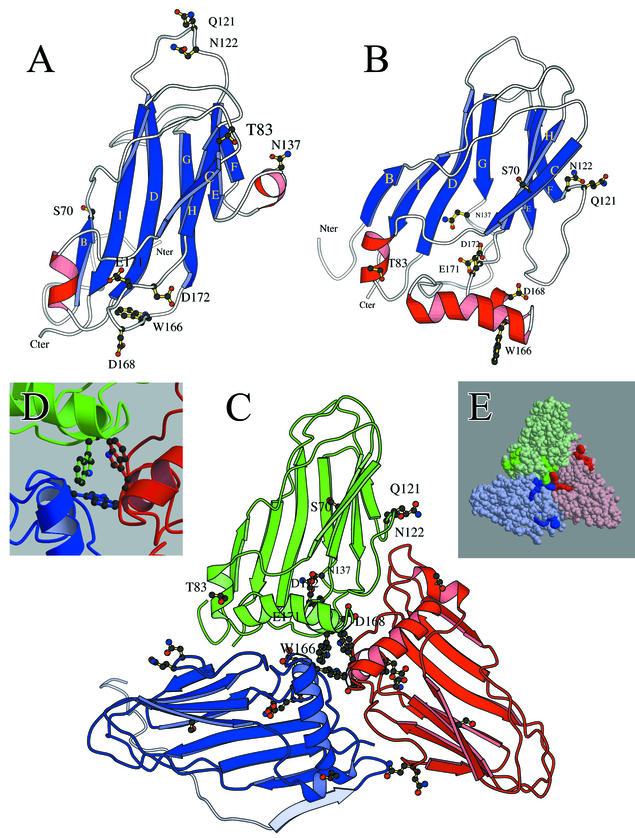
FIG.5.
(A and B) Structures of the BWYV S domain (A conformation) predicted in the Terradot model (19) (A) and in this paper (B). The views look down on the outer surface, and amino acid residues targeted for mutation in the present work are identified. The blue ribbons correspond to β-sheets, and the red coils correspond to α-helices (Fig. 4). The R domain, which is believed to extend into the capsid interior from the N terminus of the S domain (Nter), is not shown. (C) Asymmetric trimer of subunits in the A (green), B (red), and C (blue) conformations derived from our BWYV P3 S-domain structure (panel B). Amino acids targeted for mutation are identified in the green subunit. For the blue subunit, the R domain is shown in light blue. The β-sheet (light blue ribbon) near the C terminus of the R domain was predicted by computer modeling (unpublished observations), but the rest of the R domain sequence is depicted as an arbitrarily drawn disordered structure. (D) Positions of W166 residues on the subunits at the center of the trimer shown in panel C. Chains in different subunits are colored as in panel C. (E) Solid-surface representation of the structure shown in panel C. The darker colors on each subunit indicate the positions of residues corresponding to PLRV epitope 5 (the elongated structures near the sides of the triangle) and epitope 10 (the trilobate structures at the center).