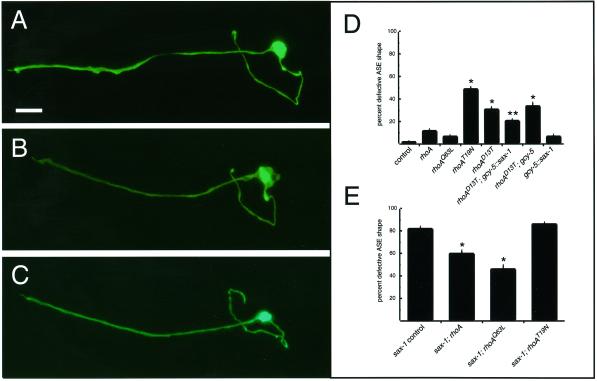
Figure 6.
ASER cell shape is altered by expression of rhoA mutant alleles. Neuronal morphology was characterized with the gcy-5::gfp transgene. All panels show a lateral view of one side of the animal. Anterior is to the left, dorsal at top. Bar, 10 μm. (A) ASER morphology in a wild-type animal. (B) Expanded cell shape defect in an animal expressing rhoAD13T in ASER. Note that this defect is milder than typical sax-1 defects (Figure 1). (C) Wild-type morphology in an animal overexpressing both rhoAD13T and sax-1 in ASER. (D and E) Effects of rhoA (wild type), rhoAQ63L (predicted gain of function), rhoAT19N (predicted dominant negative), or rhoAD13T (predicted dominant negative) expression on ASER cell shape in wild-type and mutant backgrounds. rhoA defects were weaker than those of sax-1 mutants. When scored by the criteria used for sax-1 in Figures 2, 3, and 5 (a twofold increase in cell size), the rhoA mutants did not cause significant defects (cells expressing GFP alone were 2% defective, cells expressing rhoA cDNAs were 4–8% defective). Therefore, a less stringent criterion of a 1.5-fold increase was used to score rhoA defects. By this criterion, sax-1 mutant severity did not change (compare Figure 6E to Figures 2, 3, and 5), but significant rhoA phenotypes could be detected. (D) Expression of rhoA alleles in wild-type animals. Significant cell shape defects (asterisks) were observed in ASER after expression of the predicted dominant negative rhoAT19N and rhoAD13T transgenes (single asterisks, p < 0.001, χ2 test). Overexpression of sax-1 in ASER with the use of the gcy-5 promoter partly suppressed the dominant negative effect of rhoAD13T (double asterisk, different from both control and rhoAD13T lines at p < 0.001, χ2 test). The gcy-5 promoter alone did not suppress the rhoAD13T defects. (E) Expression of rhoA alleles in sax-1(ky491) animals. The rhoA (wild type) and rhoAQ63L (predicted gain of function) alleles partly suppressed the sax-1 defect (p < 0.001, χ2 test). The predicted dominant negative rhoAT19N allele did not enhance the sax-1 defect. Control animals were injected with the coinjection marker alone. All phenotypes were scored at 25°C. Error bars indicate the SE of proportion, combining animals from four independent transgenic lines; n > 50 animals scored for each line.