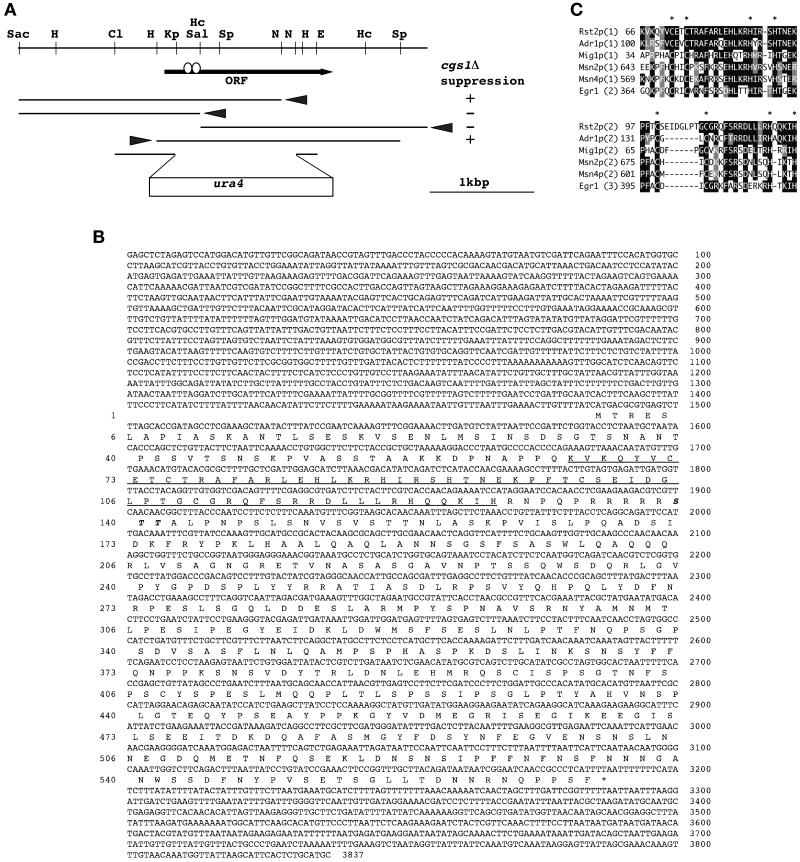
Figure 1.
The structure of the rst2 gene. (A) A restriction map of the rst2 locus and functional analysis of the subclones. The arrow indicates the position and direction of the rst2 ORF. Open circles on the arrow represent the two Cys2His2 zinc-finger motifs. Arrowheads indicate the orientation of transcription from a cryptic promoter on the vector. Each subclone was examined for the ability to promote mating and sporulation in JZ858 (cgs1Δ). The construct used to disrupt the rst2 gene is shown at the bottom. Restriction sites are abbreviated as follows: Cl, ClaI; E, EcoRI; H, HindIII; Hc, HincII; Kp, KpnI; N, NdeI; Sac, SacI; Sal, SalI; and Sp, SphI. (B) The nucleotide sequence of a 3.8-kb SacI–SphI fragment carrying rst2 and the amino acid sequence of the deduced gene product. The two zinc-finger motifs are underlined. Amino acid residues that may be phosphorylated by PKA are italicized. (C) Comparison of zinc-finger motifs among Rst2p and its close homologues. The two zinc-finger motifs of Rst2p (residues 66–128) are aligned with S. cerevisiae Adr1p (residues 100–155; Hartshorne et al., 1986), S. cerevisiae Mig1p (residues 34–90; Nehlin and Ronne, 1990), S. cerevisiae Msn2p (residues 643–698), S. cerevisiae Msn4p (residues 569–624; Estruch and Carlson, 1993), and the human EGR1 gene product (residues 364–419; Sukhatme et al., 1988). Amino acid residues identical to those of Rst2p are shown in white against black. Conserved amino acids are shown in white against gray. The number of fingers assigned for each individual protein is indicated in parentheses. Asterisks indicate cysteine and histidine residues conserved in the zinc-finger motif.