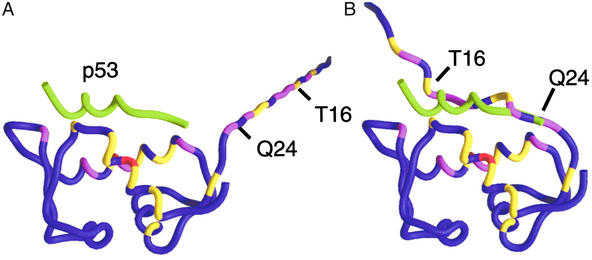
Figure 3.
Chemical shift perturbations (ΔCS) from Fig. 2 are mapped onto the Mdm2 backbone. Coloring indicates the strength of the perturbations: red, ΔCS > 0.3 ppm; magenta, 0.2 ppm < ΔCS < 0.3 ppm; and yellow, 0.1 ppm < ΔCS < 0.2 ppm. The protein used in the binding studies contained nonnative residues 9–15. Residues 9 and 10 were not assigned and were excluded. Nonnative residues 11–15 and native Mdm2 residues 16–24 were appended to the crystal structure of the p53–Mdm2 complex. (A) The chemical shift perturbations are inconsistent with a flexible, extended N terminus because, in this model, many strong perturbations occur 20–40 Å from the p53-binding site. (B) Docking the flexible Mdm2 N terminus places perturbed residues 16–24 within 5 Å of perturbed Mdm2 atoms in the p53-binding site. The Mdm2 residues 16–24 form a lid that competes weakly with p53. Perturbations to the Mdm2 residues in the p53-binding site are primarily ligand-induced; they change sign and intensity as a function of ligand composition, whereas perturbations to the lid are caused by its nonspecific displacement and are independent of the chemical nature of the ligand. The position of p53 (green tube) is included as a point of reference.