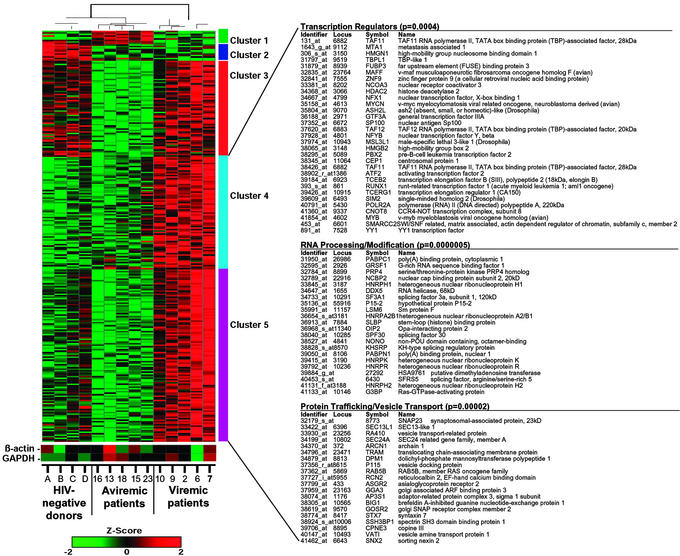
Figure 4.
Clustering of differentially expressed genes in resting CD4+ T cells of viremic patients, aviremic patients, and healthy HIV-negative donors. Transcription profiles of resting cells from five viremic patients, five aviremic patients, and four HIV-negative donors were examined by using high-density microarrays. Levels of gene expression were assayed on Affymetrix U95A chips, and 535 differentially expressed genes were identified. Genes were grouped by using K-means clustering, and samples were grouped by hierarchical clustering. Clusters 4 and 5 indicate a set of 370 genes that were up-regulated in viremic patients relative to both aviremic and healthy individuals. Statistical analysis (Fisher exact test) of the genes in clusters 4 and 5 determined that the top three functional categories, transcription regulators, RNA processing/modification, and protein trafficking/vesicle, were overrepresented in this list of genes relative to all annotated genes on the chip. Differences in relative levels of gene expression (Z-score) are indicated in color, where red indicates up-regulation and green indicates down-regulation. Some genes were listed twice because certain probes recognize multiple regions of a single gene. Levels of expression of housekeeping genes are also shown.