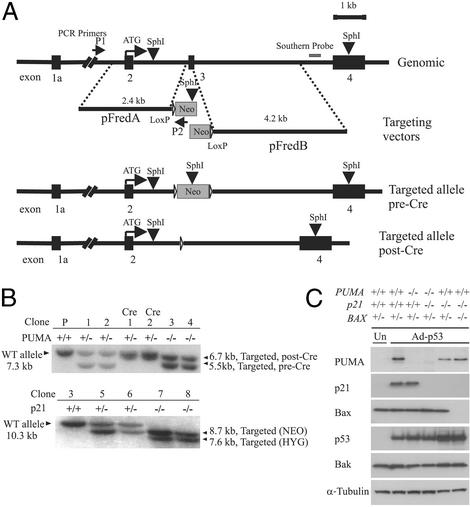
Figure 1.
Targeted deletion of PUMA. (A) Two targeting constructs were designed, each containing a homologous arm and an overlapping fragment of the neomycin-resistant gene. The SphI sites within the PUMA gene, the targeting construct, and the position of the probe used for Southern blotting are shown. PCR screening primers P1 and P2 were described in Materials and Methods. Homologous recombination results in the deletion of exon 3, which contains the BH3 domain. Filled boxes represent PUMA exons 1a–4. The same constructs were used in the second round of targeting after excision of the neomycin-resistance gene by Cre recombinase. (B Upper) A Southern blot obtained with a PUMA probe after SphI digestion of genomic DNA. (Lower) The blot obtained with a p21 probe after BglII digestion. Clone 3 (PUMA−/−, Upper) was used for targeting of the p21 locus, generating clones 5 and 6 (p21+/−). A second round of targeting generated clones 7 and 8, which were p21−/−. (C) Lysates from cells with the indicated genotypes were analyzed by immunoblotting with antibodies against PUMA, p21, p53, Bax, Bak, and α-tubulin. Cells were infected with Ad-p53 24 h before preparation of the lysates. Uninfected parental HCT116 cells (“Un”) show the basal level of p53-induced proteins in the absence of Ad-p53.